The Apaf-1*procaspase-9 apoptosome complex functions as a proteolytic-based molecular timer
- PMID: 19494828
- PMCID: PMC2711187
- DOI: 10.1038/emboj.2009.152
The Apaf-1*procaspase-9 apoptosome complex functions as a proteolytic-based molecular timer
Abstract
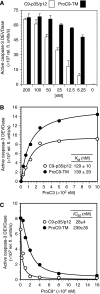
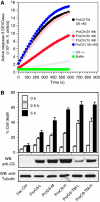
During stress-induced apoptosis, the initiator caspase-9 is activated by the Apaf-1 apoptosome and must remain bound to retain significant catalytic activity. Nevertheless, in apoptotic cells the vast majority of processed caspase-9 is paradoxically observed outside the complex. We show herein that apoptosome-mediated cleavage of procaspase-9 occurs exclusively through a CARD-displacement mechanism, so that unlike the effector procaspase-3, procaspase-9 cannot be processed by the apoptosome as a typical substrate. Indeed, procaspase-9 possessed higher affinity for the apoptosome and could displace the processed caspase-9 from the complex, thereby facilitating a continuous cycle of procaspase-9 recruitment/activation, processing, and release from the complex. Owing to its rapid autocatalytic cleavage, however, procaspase-9 per se contributed little to the activation of procaspase-3. Thus, the Apaf-1 apoptosome functions as a proteolytic-based 'molecular timer', wherein the intracellular concentration of procaspase-9 sets the overall duration of the timer, procaspase-9 autoprocessing activates the timer, and the rate at which the processed caspase-9 dissociates from the complex (and thus loses its capacity to activate procaspase-3) dictates how fast the timer 'ticks' over.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Similar articles
-
MicroRNA-17-mediated down-regulation of apoptotic protease activating factor 1 attenuates apoptosome formation and subsequent apoptosis of cardiomyocytes.Biochem Biophys Res Commun. 2015 Sep 18;465(2):299-304. doi: 10.1016/j.bbrc.2015.08.028. Epub 2015 Aug 8. Biochem Biophys Res Commun. 2015. PMID: 26265044
-
Defective molecular timer in the absence of nucleotides leads to inefficient caspase activation.PLoS One. 2011 Jan 27;6(1):e16379. doi: 10.1371/journal.pone.0016379. PLoS One. 2011. PMID: 21297999 Free PMC article.
-
Apaf-1: Regulation and function in cell death.Biochimie. 2017 Apr;135:111-125. doi: 10.1016/j.biochi.2017.02.001. Epub 2017 Feb 9. Biochimie. 2017. PMID: 28192157 Review.
-
Mechanistic insights into caspase-9 activation by the structure of the apoptosome holoenzyme.Proc Natl Acad Sci U S A. 2017 Feb 14;114(7):1542-1547. doi: 10.1073/pnas.1620626114. Epub 2017 Jan 31. Proc Natl Acad Sci U S A. 2017. PMID: 28143931 Free PMC article.
-
New insights into apoptosome structure and function.Cell Death Differ. 2018 Jul;25(7):1194-1208. doi: 10.1038/s41418-017-0025-z. Epub 2018 May 15. Cell Death Differ. 2018. PMID: 29765111 Free PMC article. Review.
Cited by
-
Up-regulation of VEGF and its receptor in refractory leukemia cells.Int J Clin Exp Pathol. 2015 May 1;8(5):5282-90. eCollection 2015. Int J Clin Exp Pathol. 2015. PMID: 26191229 Free PMC article.
-
Mitochondria and cell death: outer membrane permeabilization and beyond.Nat Rev Mol Cell Biol. 2010 Sep;11(9):621-32. doi: 10.1038/nrm2952. Epub 2010 Aug 4. Nat Rev Mol Cell Biol. 2010. PMID: 20683470 Review.
-
Cytochrome c Deficiency Confers Apoptosome and Mitochondrial Dysfunction in African-American Men with Prostate Cancer.Cancer Res. 2019 Apr 1;79(7):1353-1368. doi: 10.1158/0008-5472.CAN-18-2383. Epub 2019 Feb 14. Cancer Res. 2019. PMID: 30765600 Free PMC article.
-
Molecular mechanisms of inflammasome activation during microbial infections.Immunol Rev. 2011 Sep;243(1):174-90. doi: 10.1111/j.1600-065X.2011.01041.x. Immunol Rev. 2011. PMID: 21884176 Free PMC article. Review.
-
Cdc6 protein obstructs apoptosome assembly and consequent cell death by forming stable complexes with activated Apaf-1 molecules.J Biol Chem. 2012 May 25;287(22):18573-83. doi: 10.1074/jbc.M112.347690. Epub 2012 Apr 6. J Biol Chem. 2012. PMID: 22493447 Free PMC article.
References
-
- Acehan D, Jiang X, Morgon DG, Heuser JE, Wang X, Akey CW (2002) Three-dimensional structure of the apoptosome: implications for assembly, procaspase-9 binding, and activation. Mol Cell 9: 423–432 - PubMed
-
- Allan LA, Clarke PR (2007) Phosphorylation of caspase-9 by CDK1/cyclin B1 protects mitotic cells against apoptosis. Mol Cell 26: 301–310 - PubMed
-
- Allan LA, Morrice N, Brady S, Magee G, Pathak S, Clarke PR (2003) Inhibition of caspase-9 through phosphorylation at Thr 125 by ERK MAPK. Nat Cell Biol 5: 647–654 - PubMed
-
- Benedict MA, Hu Y, Inohara N, Nunez G (2000) Expression and functional analysis of Apaf-1 isoforms. Extra WD-40 repeat is required for cytochrome c binding and regulated activation of procaspase-9. J Biol Chem 275: 8461–8468 - PubMed
-
- Boatright KM, Renatus M, Scott FL, Sperandio S, Shin H, Pedersen IM, Ricci JE, Edris WA, Sutherlin DP, Green DR, Salvesen GS (2003) A unified model for apical caspase activation. Mol Cell 11: 529–541 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

