Genomic characterization of methanomicrobiales reveals three classes of methanogens
- PMID: 19495416
- PMCID: PMC2686161
- DOI: 10.1371/journal.pone.0005797
Genomic characterization of methanomicrobiales reveals three classes of methanogens
Abstract
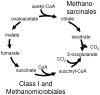
Background: Methanomicrobiales is the least studied order of methanogens. While these organisms appear to be more closely related to the Methanosarcinales in ribosomal-based phylogenetic analyses, they are metabolically more similar to Class I methanogens.
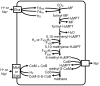
Methodology/principal findings: In order to improve our understanding of this lineage, we have completely sequenced the genomes of two members of this order, Methanocorpusculum labreanum Z and Methanoculleus marisnigri JR1, and compared them with the genome of a third, Methanospirillum hungatei JF-1. Similar to Class I methanogens, Methanomicrobiales use a partial reductive citric acid cycle for 2-oxoglutarate biosynthesis, and they have the Eha energy-converting hydrogenase. In common with Methanosarcinales, Methanomicrobiales possess the Ech hydrogenase and at least some of them may couple formylmethanofuran formation and heterodisulfide reduction to transmembrane ion gradients. Uniquely, M. labreanum and M. hungatei contain hydrogenases similar to the Pyrococcus furiosus Mbh hydrogenase, and all three Methanomicrobiales have anti-sigma factor and anti-anti-sigma factor regulatory proteins not found in other methanogens. Phylogenetic analysis based on seven core proteins of methanogenesis and cofactor biosynthesis places the Methanomicrobiales equidistant from Class I methanogens and Methanosarcinales.
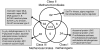
Conclusions/significance: Our results indicate that Methanomicrobiales, rather than being similar to Class I methanogens or Methanomicrobiales, share some features of both and have some unique properties. We find that there are three distinct classes of methanogens: the Class I methanogens, the Methanomicrobiales (Class II), and the Methanosarcinales (Class III).
Conflict of interest statement
Figures

References
-
- Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. Mesophilic crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol. 2008;6:245–252. - PubMed
-
- Ferry JG. Methane: small molecule, big impact. Science. 1997;278:1413–1414. - PubMed
-
- Etheridge DM, Steele LP, Francey RJ, Langenfelds RL. Atmospheric methane between 1000 A.D. and present: evidence of anthropogenic emissions and climatic variability. J Geophys Res. 1998;103:15979–15993.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

