Mechanism of strand-specific smooth muscle alpha-actin enhancer interaction by purine-rich element binding protein B (Purbeta)
- PMID: 19496623
- PMCID: PMC2752054
- DOI: 10.1021/bi900708j
Mechanism of strand-specific smooth muscle alpha-actin enhancer interaction by purine-rich element binding protein B (Purbeta)
Abstract
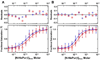
Expression of the smooth muscle alpha-actin gene in growth-activated vascular smooth muscle cells and stromal fibroblasts is negatively regulated by members of the Pur family of single-stranded DNA/RNA-binding proteins. In particular, Puralpha and Purbeta are postulated to repress transcription by forming helix-destabilizing complexes with the sense strand of an asymmetric polypurine-polypyrimidine tract containing a canonical MCAT enhancer motif in the 5' region of the gene. Herein, we establish the mechanism of Purbeta binding to the purine-rich strand of the enhancer using quantitative methods and purified components. Initial evaluation of DNA-binding specificity and equilibrium stoichiometry via colorimetric-, autoradiographic-, and fluorescence-based assays suggested that Purbeta interacts with two distinct G/A-rich sites within the nominal single-stranded enhancer element to form a high-affinity 2:1 protein:DNA complex. Statistical mechanical analyses of band shift titrations of the nominal element in conjunction with DNase I footprint titrations of the extended smooth muscle alpha-actin 5'-flanking region demonstrated that assembly of the nucleoprotein complex likely occurs in a sequential, cooperative, and monomer-dependent fashion. Resolution of the microscopic energetics of the system indicated that monomer association with two nonidentical sites flanking the core MCAT motif accounts for the majority of the intrinsic binding affinity of Purbeta with intersite cooperativity contributing an approximately 12-fold increase to the stability of the nucleoprotein complex. These findings offer new insights into the mechanism, energetics, and sequence determinants of Purbeta repressor binding to a biologically relevant, contractile phenotype-regulating cis-element while also revealing the thermodynamic confines of putative Purbeta-mediated effects on DNA structure.
Figures






References
-
- Wang ZY, Lin XH, Nobyuoshi M, Qui QQ, Deuel TF. Binding of single-stranded oligonucleotides to a non-B-form DNA structure results in loss of promoter activity of the platelet-derived growth factor A-chain gene. J. Biol. Chem. 1992;267:13669–13674. - PubMed
-
- Chen S, Supakar PC, Vellanoweth RL, Song CS, Chatterjee B, Roy AK. Functional role of a conformationally flexible homopurine/homopyrimidine domain of the androgen receptor gene promoter interacting with Sp1 and a pyrimidine single strand DNA-binding protein. Mol. Endocrinol. 1997;11:3–15. - PubMed
-
- Rustighi A, Tessari MA, Vascotto F, Sgarra R, Giancotti V, Manfioletti G. A polypyrimidine/polypurine tract within the Hmga2 minimal promoter: a common feature of many growth-related genes. Biochemistry. 2002;41:1229–1240. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

