Intracellular interactions between APOBEC3G, RNA, and HIV-1 Gag: APOBEC3G multimerization is dependent on its association with RNA
- PMID: 19497112
- PMCID: PMC2700067
- DOI: 10.1186/1742-4690-6-56
Intracellular interactions between APOBEC3G, RNA, and HIV-1 Gag: APOBEC3G multimerization is dependent on its association with RNA
Abstract
Background: Host restriction factor APOBEC3G (A3G) blocks human immunodeficiency virus type 1 (HIV-1) replication by G-to-A hypermutation, and by inhibiting DNA synthesis and provirus formation. Previous reports have suggested that A3G is a dimer and its virion incorporation is mediated through interactions with viral or nonviral RNAs and/or HIV-1 Gag. We have now employed a bimolecular fluorescence complementation assay (BiFC) to analyze the intracellular A3G-A3G, A3G-RNA, and A3G-Gag interactions in living cells by reconstitution of yellow fluorescent protein (YFP) from its N- or C-terminal fragments.
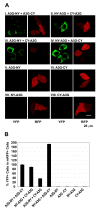
Results: The results obtained with catalytic domain 1 and 2 (CD1 and CD2) mutants indicate that A3G-A3G and A3G-Gag multimerization is dependent on an intact CD1 domain, which is required for RNA binding. A mutant HIV-1 Gag that exhibits reduced RNA binding also failed to reconstitute BiFC with wild-type A3G, indicating a requirement for both HIV-1 Gag and A3G to bind to RNA for their multimerization. Addition of a non-specific RNA binding peptide (P22) to the N-terminus of a CD1 mutant of A3G restored BiFC and virion incorporation, but failed to inhibit viral replication, indicating that the mutations in CD1 resulted in additional defects that interfere with A3G's antiviral activity.
Conclusion: These studies establish a robust BiFC assay for analysis of intracellular interactions of A3G with other macromolecules. The results indicate that in vivo A3G is a monomer that forms multimers upon binding to RNA. In addition, we observed weak interactions between wild-type A3G molecules and RNA binding-defective mutants of A3G, which could explain previously described protein-protein interactions between purified A3G molecules.
Figures








Similar articles
-
Mechanism of Enhanced HIV Restriction by Virion Coencapsidated Cytidine Deaminases APOBEC3F and APOBEC3G.J Virol. 2017 Jan 18;91(3):e02230-16. doi: 10.1128/JVI.02230-16. Print 2017 Feb 1. J Virol. 2017. PMID: 27881650 Free PMC article.
-
Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs.J Biol Chem. 2004 Aug 20;279(34):35822-8. doi: 10.1074/jbc.M405761200. Epub 2004 Jun 20. J Biol Chem. 2004. PMID: 15210704
-
APOBEC3G induces a hypermutation gradient: purifying selection at multiple steps during HIV-1 replication results in levels of G-to-A mutations that are high in DNA, intermediate in cellular viral RNA, and low in virion RNA.Retrovirology. 2009 Feb 13;6:16. doi: 10.1186/1742-4690-6-16. Retrovirology. 2009. PMID: 19216784 Free PMC article.
-
Apobec3G-Based Strategies to Defeat HIV Infection.Curr HIV Res. 2016;14(3):217-24. doi: 10.2174/1570162x14999160224100541. Curr HIV Res. 2016. PMID: 26957196 Review.
-
[Advances in the study of molecular mechanism of APOBEC3G anti-HIV-1].Yao Xue Xue Bao. 2008 Jul;43(7):678-82. Yao Xue Xue Bao. 2008. PMID: 18819469 Review. Chinese.
Cited by
-
Identification of a dominant negative inhibitor of human zinc finger antiviral protein reveals a functional endogenous pool and critical homotypic interactions.J Virol. 2010 May;84(9):4504-12. doi: 10.1128/JVI.02018-09. Epub 2010 Feb 24. J Virol. 2010. PMID: 20181706 Free PMC article.
-
Design of Vif-Derived Peptide Inhibitors with Anti-HIV-1 Activity by Interrupting Vif-CBFβ Interaction.Viruses. 2024 Mar 22;16(4):490. doi: 10.3390/v16040490. Viruses. 2024. PMID: 38675833 Free PMC article.
-
Sequence and structural determinants of human APOBEC3H deaminase and anti-HIV-1 activities.Retrovirology. 2015 Jan 22;12:3. doi: 10.1186/s12977-014-0130-8. Retrovirology. 2015. PMID: 25614027 Free PMC article.
-
P body-associated protein Mov10 inhibits HIV-1 replication at multiple stages.J Virol. 2010 Oct;84(19):10241-53. doi: 10.1128/JVI.00585-10. Epub 2010 Jul 28. J Virol. 2010. PMID: 20668078 Free PMC article.
-
Modeling the Embrace of a Mutator: APOBEC Selection of Nucleic Acid Ligands.Trends Biochem Sci. 2018 Aug;43(8):606-622. doi: 10.1016/j.tibs.2018.04.013. Epub 2018 May 23. Trends Biochem Sci. 2018. PMID: 29803538 Free PMC article. Review.
References
-
- Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. - PubMed
-
- Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. - PubMed
-
- Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. - PubMed
-
- Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. - PubMed
-
- Lecossier D, Bouchonnet F, Clavel F, Hance AJ. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science. 2003;300:1112. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

