Dynamic positional fate map of the primary heart-forming region
- PMID: 19497319
- PMCID: PMC2720636
- DOI: 10.1016/j.ydbio.2009.05.570
Dynamic positional fate map of the primary heart-forming region
Abstract
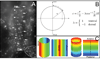
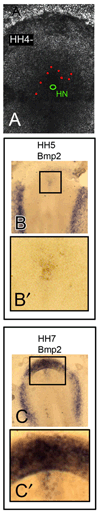
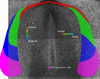
Here we show the temporal-spatial orchestration of early heart morphogenesis at cellular level resolution, in vivo, and reconcile conflicting positional fate mapping data regarding the primary heart-forming field(s). We determined the positional fates of precardiac cells using a precision electroporation approach in combination with wide-field time-lapse microscopy in the quail embryo, a warm-blooded vertebrate (HH Stages 4 through 10). Contrary to previous studies, the results demonstrate the existence of a "continuous" circle-shaped heart field that spans the midline, appearing at HH Stage 4, which then expands to form a wide arc of progenitors at HH Stages 5-7. Our time-resolved image data show that a subset of these cardiac progenitor cells do not overlap with the expression of common cardiogenic factors, Nkx-2.5 and Bmp-2, until HH Stage 10, when a tubular heart has formed, calling into question when cardiac fate is specified and by which key factors. Sub-groups and anatomical bands (cohorts) of heart precursor cells dramatically change their relative positions in a process largely driven by endodermal folding and other large-scale tissue deformations. Thus, our novel dynamic positional fate maps resolve the origin of cardiac progenitor cells in amniotes. The data also establish the concept that tissue motion contributes significantly to cellular position fate - i.e., much of the cellular displacement that occurs during assembly of a midline heart tube (HH Stage 9) is NOT due to "migration" (autonomous motility), a commonly held belief. Computational analysis of our time-resolved data lays the foundation for more precise analyses of how cardiac gene regulatory networks correlate with early heart tissue morphogenesis in birds and mammals.
Figures




References
-
- Abu-Issa R, Kirby ML. Heart Field: From Mesoderm to Heart Tube. Annu Rev Cell Dev Biol. 2007 - PubMed
-
- Abu-Issa R, Waldo K, Kirby ML. Heart fields: one, two or more? Dev Biol. 2004;272:281–285. - PubMed
-
- Baldwin HS, Jensen KL, Solursh M. Myogenic cytodifferentiation of the precardiac mesoderm in the rat. Differentiation. 1991;47:163–172. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

