Formyl peptide receptors are candidate chemosensory receptors in the vomeronasal organ
- PMID: 19497865
- PMCID: PMC2690606
- DOI: 10.1073/pnas.0904464106
Formyl peptide receptors are candidate chemosensory receptors in the vomeronasal organ
Abstract
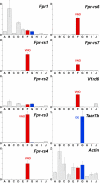
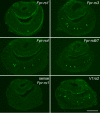
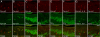
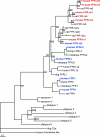
The identification of receptors that detect environmental stimuli lays a foundation for exploring the mechanisms and neural circuits underlying sensation. The mouse vomeronasal organ (VNO), which detects pheromones and other semiochemicals, has 2 known families of chemoreceptors, V1Rs and V2Rs. Here, we report a third family of mouse VNO receptors comprising 5 of 7 members of the formyl peptide receptor (FPR) family. Unlike other FPRs, which function in the immune system, these FPRs are selectively expressed in VNO neurons in patterns strikingly similar to those of V1Rs and V2Rs. Each FPR is expressed in a different small subset of neurons that are highly dispersed in the neuroepithelium, consistently coexpress either G alpha(i2) or G alpha(o), and lack other chemoreceptors examined. Given the presence of formylated peptides in bacteria and mitochondria, possible roles for VNO FPRs include the assessment of conspecifics or other species based on variations in normal bacterial flora or mitochondrial proteins.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Brennan PA, Keverne EB. Something in the air? New insights into mammalian pheromones. Curr Biol. 2004;14(2):R81–R89. - PubMed
-
- Dulac C, Torello AT. Molecular detection of pheromone signals in mammals: from genes to behaviour. Nat Rev Neurosci. 2003;4(7):551–562. - PubMed
-
- Halpern M, Martinez-Marcos A. Structure and function of the vomeronasal system: an update. Progress in neurobiology. 2003;70(3):245–318. - PubMed
-
- Kandel ER, Schwartz JH, Jessell TM. Principles of neural science. New York: McGraw-Hill; 2000. pp. 625–647.
-
- Boehm T, Zufall F. MHC peptides and the sensory evaluation of genotype. Trends in neurosciences. 2006;29(2):100–107. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

