Dynamic histone H3 epigenome marking during the intraerythrocytic cycle of Plasmodium falciparum
- PMID: 19497874
- PMCID: PMC2701018
- DOI: 10.1073/pnas.0902515106
Dynamic histone H3 epigenome marking during the intraerythrocytic cycle of Plasmodium falciparum
Abstract
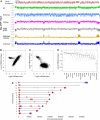
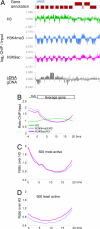
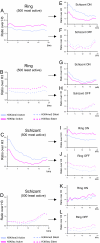
Epigenome profiling has led to the paradigm that promoters of active genes are decorated with H3K4me3 and H3K9ac marks. To explore the epigenome of Plasmodium falciparum asexual stages, we performed MS analysis of histone modifications and found a general preponderance of H3/H4 acetylation and H3K4me3. ChIP-on-chip profiling of H3, H3K4me3, H3K9me3, and H3K9ac from asynchronous parasites revealed an extensively euchromatic epigenome with heterochromatin restricted to variant surface antigen gene families (VSA) and a number of genes hitherto unlinked to VSA. Remarkably, the vast majority of the genome shows an unexpected pattern of enrichment of H3K4me3 and H3K9ac. Analysis of synchronized parasites revealed significant developmental stage specificity of the epigenome. In rings, H3K4me3 and H3K9ac are homogenous across the genes marking active and inactive genes equally, whereas in schizonts, they are enriched at the 5' end of active genes. This study reveals an unforeseen and unique plasticity in the use of the epigenetic marks and implies the presence of distinct epigenetic pathways in gene silencing/activation throughout the erythrocytic cycle.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Arnot DE, Gull K. The Plasmodium cell-cycle: Facts and questions. Ann Trop Med Parasitol. 1998;92:361–365. - PubMed
-
- Le Roch KG, et al. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301:1503–1508. - PubMed
-
- Florens L, et al. A proteomic view of the Plasmodium falciparum life cycle. Nature. 2002;419:520–526. - PubMed
-
- Lasonder E, et al. Analysis of the Plasmodium falciparum proteome by high-accuracy mass spectrometry. Nature. 2002;419:537–542. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

