Seeing the forest for the trees: using the Gene Ontology to restructure hierarchical clustering
- PMID: 19497934
- PMCID: PMC2705235
- DOI: 10.1093/bioinformatics/btp327
Seeing the forest for the trees: using the Gene Ontology to restructure hierarchical clustering
Abstract
Motivation: There is a growing interest in improving the cluster analysis of expression data by incorporating into it prior knowledge, such as the Gene Ontology (GO) annotations of genes, in order to improve the biological relevance of the clusters that are subjected to subsequent scrutiny. The structure of the GO is another source of background knowledge that can be exploited through the use of semantic similarity.
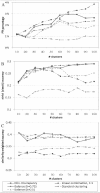
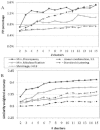
Results: We propose here a novel algorithm that integrates semantic similarities (derived from the ontology structure) into the procedure of deriving clusters from the dendrogram constructed during expression-based hierarchical clustering. Our approach can handle the multiple annotations, from different levels of the GO hierarchy, which most genes have. Moreover, it treats annotated and unannotated genes in a uniform manner. Consequently, the clusters obtained by our algorithm are characterized by significantly enriched annotations. In both cross-validation tests and when using an external index such as protein-protein interactions, our algorithm performs better than previous approaches. When applied to human cancer expression data, our algorithm identifies, among others, clusters of genes related to immune response and glucose metabolism. These clusters are also supported by protein-protein interaction data.
Figures
References
-
- Bellazzi R, Zupan B. Towards knowledge-based gene expression data mining. J. Biomed. Inform. 2007;6:787–802. - PubMed
-
- Buehler EC, et al. The CRASSS plug-in for integrating annotation data with hierarchical clustering results. Bioinformatics. 2004;20:3266–3269. - PubMed
-
- Cheng J, et al. A knowledge-based clustering algorithm driven by Gene Ontology. J. Biopharm. Stat. 2004;14:687–700. - PubMed
-
- Crocker PR, et al. Siglecs and their roles in the immune system. Nat. Rev. Immunol. 2007;7:255–266. - PubMed
-
- Curtis RK, et al. Pathways to the analysis of microarray data. Trends Biotechnol. 2005;23:429–435. - PubMed

