Histone-acetylated control of fibroblast growth factor receptor 2 intron 2 polymorphisms and isoform splicing in breast cancer
- PMID: 19497954
- PMCID: PMC2737561
- DOI: 10.1210/me.2009-0071
Histone-acetylated control of fibroblast growth factor receptor 2 intron 2 polymorphisms and isoform splicing in breast cancer
Abstract
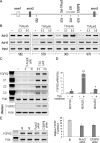
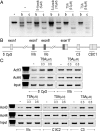
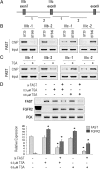
Recent genome-wide association studies have identified fibroblast growth factor receptor (FGFR)2 as one of a few candidate genes linked with breast cancer susceptibility. In particular, the disease-predisposing allele of FGFR2 is inherited as a 7.5-kb region within intron 2 that harbors eight single nucleotide polymorphisms. The relationship between these single nucleotide polymorphisms and FGFR2 gene expression remains unclear. Here we show the common occurrence of polymorphisms within the intron 2 region in a panel of 10 breast cancer cell lines. High FGFR2-expressing cell lines such as MCF-7 cells displayed polymorphic sequences with constitutive histone acetylation at multiple intron 2 sequences harboring putative transcription binding sites. Knockdown of Runx2 or CCAAT enhancer binding protein beta in these cells resulted in diminished endogenous FGFR2 gene expression. In contrast FGFR2-negative MDA-231 cells were wild type and showed evidence of histone 3/4 deacetylation at the rs2981578, rs10736303, and rs7895676 disease-associated alleles that harbor binding sites for Runx2, estrogen receptor, and CCAAT enhancer binding protein beta, respectively. Histone deacetylation inhibition with trichostatin A resulted in enhanced acetylation at these intron 2 sites, an effect associated with robust FGFR2 reexpression. Isoform analysis proved reexpression of the FGFR2-IIIc variant the splicing of which was positively influenced by trichostatin A-mediated recruitment of the Fas-activated serine/threonine phosphoprotein survival protein. Our findings highlight the potential role of histone acetylation in modulating access to selected polymorphic sites within intron 2 as well as downstream splicing sites in generating variable FGFR2 levels and isoforms in breast cancer.
Figures


Similar articles
-
Functional analysis of a breast cancer-associated FGFR2 single nucleotide polymorphism using zinc finger mediated genome editing.PLoS One. 2013 Nov 12;8(11):e78839. doi: 10.1371/journal.pone.0078839. eCollection 2013. PLoS One. 2013. PMID: 24265722 Free PMC article.
-
Genetic and epigenetic mechanisms down-regulate FGF receptor 2 to induce melanoma-associated antigen A in breast cancer.Am J Pathol. 2010 May;176(5):2333-43. doi: 10.2353/ajpath.2010.091049. Epub 2010 Mar 26. Am J Pathol. 2010. Retraction in: Am J Pathol. 2015 Aug;185(8):2339. doi: 10.1016/j.ajpath.2015.04.025. PMID: 20348248 Free PMC article. Retracted.
-
A study on genetic variants of Fibroblast growth factor receptor 2 (FGFR2) and the risk of breast cancer from North India.PLoS One. 2014 Oct 21;9(10):e110426. doi: 10.1371/journal.pone.0110426. eCollection 2014. PLoS One. 2014. PMID: 25333473 Free PMC article.
-
Association between FGFR2 (rs2981582, rs2420946 and rs2981578) polymorphism and breast cancer susceptibility: a meta-analysis.Oncotarget. 2017 Jan 10;8(2):3454-3470. doi: 10.18632/oncotarget.13839. Oncotarget. 2017. PMID: 27966449 Free PMC article.
-
Variants of FGFR2 and their associations with breast cancer risk: a HUGE systematic review and meta-analysis.Breast Cancer Res Treat. 2016 Jan;155(2):313-35. doi: 10.1007/s10549-015-3670-2. Epub 2016 Jan 4. Breast Cancer Res Treat. 2016. PMID: 26728143
Cited by
-
Acetylation-Mediated Epigenetic Consequences for Biological Control and Cancer.Results Probl Cell Differ. 2025;75:25-69. doi: 10.1007/978-3-031-91459-1_2. Results Probl Cell Differ. 2025. PMID: 40593206 Review.
-
Association of FGF Receptor-2rs2981578 SNP with breast cancer in women at the University of Gondar Comprehensive Specialized Hospital, Ethiopia.PLoS One. 2025 Aug 1;20(8):e0327034. doi: 10.1371/journal.pone.0327034. eCollection 2025. PLoS One. 2025. PMID: 40748883 Free PMC article.
-
Fine-scale mapping of the FGFR2 breast cancer risk locus: putative functional variants differentially bind FOXA1 and E2F1.Am J Hum Genet. 2013 Dec 5;93(6):1046-60. doi: 10.1016/j.ajhg.2013.10.026. Epub 2013 Nov 27. Am J Hum Genet. 2013. PMID: 24290378 Free PMC article.
-
CREB1K292 and HINFPK330 as Putative Common Therapeutic Targets in Alzheimer's and Parkinson's Disease.ACS Omega. 2021 Dec 16;6(51):35780-35798. doi: 10.1021/acsomega.1c05827. eCollection 2021 Dec 28. ACS Omega. 2021. PMID: 34984308 Free PMC article.
-
Fast kinase domain-containing protein 3 is a mitochondrial protein essential for cellular respiration.Biochem Biophys Res Commun. 2010 Oct 22;401(3):440-6. doi: 10.1016/j.bbrc.2010.09.075. Epub 2010 Sep 24. Biochem Biophys Res Commun. 2010. PMID: 20869947 Free PMC article.
References
-
- Givol D, Yayon A 1992 Complexity of FGF receptors: genetic basis for structural diversity and functional specificity. FASEB J 6:3362–3369 - PubMed
-
- Yan G, Wang F, Fukabori Y, Sussman D, Hou J, McKeehan WL 1992 Expression and transformation of a variant of the heparin-binding fibroblast growth factor receptor (flg) gene resulting from splicing of the exon at alternate 3′-acceptor site. Biochem Biophys Res Commun 183:423–430 - PubMed
-
- Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, Struewing JP, Morrison J, Field H, Luben R, Wareham N, Ahmed S, Healey CS, Bowman R, Meyer KB, Haiman CA, Kolonel LK, Henderson BE, Le Marchand L, Brennan P, Sangrajrang S, Gaborieau V, Odefrey F, Shen CY, Wu PE, et al. 2007 Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 447:1087–1093 - PMC - PubMed
-
- Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, Wacholder S, Wang Z, Welch R, Hutchinson A, Wang J, Yu K, Chatterjee N, Orr N, Willett WC, Colditz GA, Ziegler RG, Berg CD, Buys SS, McCarty CA, Feigelson HS, Calle EE, Thun MJ, Hayes RB, Tucker M, et al. 2007 A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet 39:870–874 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

