Development of a malignancy-associated proteomic signature for diffuse large B-cell lymphoma
- PMID: 19498000
- PMCID: PMC2708791
- DOI: 10.2353/ajpath.2009.080707
Development of a malignancy-associated proteomic signature for diffuse large B-cell lymphoma
Abstract
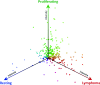
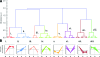
The extreme pathological diversity of non-Hodgkin's lymphomas has made their accurate histological assessment difficult. New diagnostics and treatment modalities are urgently needed for these lymphomas, particularly in drug development for cancer-specific targets. Previously, we showed that a subset of B cell lymphoma, diffuse large B cell lymphoma, may be characterized by two major, orthogonal axes of gene expression: one set of transcripts that is differentially expressed between resting and proliferating, nonmalignant cells (ie, a "proliferative signature") and another set that is expressed only in proliferating malignant cells (ie, a "cancer signature"). A differential proteomic analysis of B cell proliferative states, similar to previous transcriptional profiling analyses, holds great promise either to reveal novel factors that participate in lymphomagenesis or to define biomarkers of onset or progression. Here, we use a murine model of diffuse large B cell lymphoma to conduct unbiased two-dimensional gel electrophoresis and mass spectrometry-based comparative proteomic analyses of malignant proliferating B cells and tissue-matched, normal resting, or normal proliferating cells. We show that the expression patterns of particular proteins or isoforms across these states fall into eight specific trends that provide a framework to identify malignancy-associated biomarkers and potential drug targets, a signature proteome. Our results support the central hypothesis that clusters of proteins of known function represent a panel of expression markers uniquely associated with malignancy and not normal proliferation.
Figures



References
-
- Edwards BK, Brown ML, Wingo PA, Howe HL, Ward E, Ries LAG, Schrag D, Jamison PM, Jemal A, Wu XC, Friedman C, Harlan L, Warren J, Anderson RN, Pickle LW. Annual report to the nation on the status of cancer, 1975–2002, featuring population-based trends in cancer treatment. J Natl Cancer Inst. 2005;97:1407–1427. - PubMed
-
- Ng AK. Diffuse large B cell lymphoma. Semin Radiat Oncol. 2007;17:169–175. - PubMed
-
- Illidge T, Tolan S. Current treatment approaches for diffuse large B cell lymphoma. Leukemia Lymphoma. 2008;49:663–676. - PubMed
-
- Alizadeh AA, Staudt LM. Genomic-scale gene expression profiling of normal and malignant immune cells. Curr Opin Immunol. 2000;12:219–225. - PubMed
-
- Alizadeh AA, Eisen M, Davis RE, C Ma, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson J, Jr, Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Levy R, Wilson W, Grever MR, Byrd JC, Botstein D, Brown PO, Staudt LM. Distinct types of diffuse large-B cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

