Combining use of a panel of ssDNA aptamers in the detection of Staphylococcus aureus
- PMID: 19498077
- PMCID: PMC2724295
- DOI: 10.1093/nar/gkp489
Combining use of a panel of ssDNA aptamers in the detection of Staphylococcus aureus
Abstract
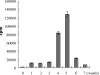
In this article, a panel of ssDNA aptamers specific to Staphylococcus aureus was obtained by a whole bacterium-based SELEX procedure and applied to probing S. aureus. After several rounds of selection with S. aureus as the target and Streptococcus and S. epidermidis as counter targets, the highly enriched oligonucleic acid pool was sequenced and then grouped under different families on the basis of the homology of the primary sequence and the similarity of the secondary structure. Eleven sequences from different families were selected for further characterization by confocal imaging and flow cytometry analysis. Results showed that five aptamers demonstrated high specificity and affinity to S. aureus individually. The five aptamers recognize different molecular targets by competitive experiment. Combining these five aptamers had a much better effect than the individual aptamer in the recognition of different S. aureus strains. In addition, the combined aptamers can probe single S. aureus in pyogenic fluids. Our work demonstrates that a set of aptamers specific to one bacterium can be used in combination for the identification of the bacterium instead of a single aptamer.
Figures



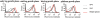

Similar articles
-
Capture and detection of Staphylococcus aureus with dual labeled aptamers to cell surface components.Int J Food Microbiol. 2018 Jan 16;265:74-83. doi: 10.1016/j.ijfoodmicro.2017.11.002. Epub 2017 Nov 7. Int J Food Microbiol. 2018. PMID: 29132030
-
Identification of Salmonella Typhimurium-specific DNA aptamers developed using whole-cell SELEX and FACS analysis.J Microbiol Methods. 2013 Nov;95(2):162-6. doi: 10.1016/j.mimet.2013.08.005. Epub 2013 Aug 24. J Microbiol Methods. 2013. PMID: 23978634
-
Isolation of a new ssDNA aptamer against staphylococcal enterotoxin B based on CNBr-activated sepharose-4B affinity chromatography.J Mol Recognit. 2016 Sep;29(9):436-45. doi: 10.1002/jmr.2542. Epub 2016 Apr 19. J Mol Recognit. 2016. PMID: 27091327
-
Recent Progress in the Identification of Aptamers Against Bacterial Origins and Their Diagnostic Applications.Int J Mol Sci. 2020 Jul 18;21(14):5074. doi: 10.3390/ijms21145074. Int J Mol Sci. 2020. PMID: 32708376 Free PMC article. Review.
-
Development of Cell-Specific Aptamers: Recent Advances and Insight into the Selection Procedures.Molecules. 2017 Nov 27;22(12):2070. doi: 10.3390/molecules22122070. Molecules. 2017. PMID: 29186905 Free PMC article. Review.
Cited by
-
DNA aptamers against the Lup an 1 food allergen.PLoS One. 2012;7(4):e35253. doi: 10.1371/journal.pone.0035253. Epub 2012 Apr 17. PLoS One. 2012. PMID: 22529997 Free PMC article.
-
Rational Design of a User-Friendly Aptamer/Peptide-Based Device for the Detection of Staphylococcus aureus.Sensors (Basel). 2020 Sep 2;20(17):4977. doi: 10.3390/s20174977. Sensors (Basel). 2020. PMID: 32887407 Free PMC article.
-
Isolation of an Aptamer that Binds Specifically to E. coli.PLoS One. 2016 Apr 22;11(4):e0153637. doi: 10.1371/journal.pone.0153637. eCollection 2016. PLoS One. 2016. PMID: 27104834 Free PMC article.
-
Advances in Immunomodulation and Immune Engineering Approaches to Improve Healing of Extremity Wounds.Int J Mol Sci. 2022 Apr 7;23(8):4074. doi: 10.3390/ijms23084074. Int J Mol Sci. 2022. PMID: 35456892 Free PMC article. Review.
-
A PDMS/paper/glass hybrid microfluidic biochip integrated with aptamer-functionalized graphene oxide nano-biosensors for one-step multiplexed pathogen detection.Lab Chip. 2013 Oct 7;13(19):3921-8. doi: 10.1039/c3lc50654a. Lab Chip. 2013. PMID: 23929394 Free PMC article.
References
-
- Lowy FD. Staphylococcus aureus infections. New Engl. J. Med. 1998;339:520–532. - PubMed
-
- Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. - PubMed
-
- Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. - PubMed
-
- Ulrich H, Magdesian MH, Alves MJ, Colli W. In vitro selection of RNA aptamers that bind to cell adhesion receptors of Trypanosoma cruzi and inhibit cell invasion. J. Biol. Chem. 2002;277:20756–20762. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

