Mechanoenzymatic cleavage of the ultralarge vascular protein von Willebrand factor
- PMID: 19498171
- PMCID: PMC2753189
- DOI: 10.1126/science.1170905
Mechanoenzymatic cleavage of the ultralarge vascular protein von Willebrand factor
Abstract
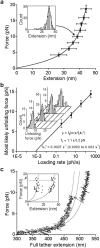
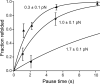
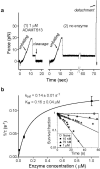
Von Willebrand factor (VWF) is secreted as ultralarge multimers that are cleaved in the A2 domain by the metalloprotease ADAMTS13 to give smaller multimers. Cleaved VWF is activated by hydrodynamic forces found in arteriolar bleeding to promote hemostasis, whereas uncleaved VWF is activated at lower, physiologic shear stresses and causes thrombosis. Single-molecule experiments demonstrate that elongational forces in the range experienced by VWF in the vasculature unfold the A2 domain, and only the unfolded A2 domain is cleaved by ADAMTS13. In shear flow, tensile force on a VWF multimer increases with the square of multimer length and is highest at the middle, providing an efficient mechanism for homeostatic regulation of VWF size distribution by force-induced A2 unfolding and cleavage by ADAMTS13, as well as providing a counterbalance for VWF-mediated platelet aggregation.
Figures


Comment in
-
Biochemistry. Force signaling in biology.Science. 2009 Jun 5;324(5932):1278-80. doi: 10.1126/science.1175874. Science. 2009. PMID: 19498156 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

