Cre-lox univector acceptor vectors for functional screening in protoplasts: analysis of Arabidopsis donor cDNAs encoding ABSCISIC ACID INSENSITIVE1-like protein phosphatases
- PMID: 19499346
- PMCID: PMC2755202
- DOI: 10.1007/s11103-009-9502-1
Cre-lox univector acceptor vectors for functional screening in protoplasts: analysis of Arabidopsis donor cDNAs encoding ABSCISIC ACID INSENSITIVE1-like protein phosphatases
Abstract
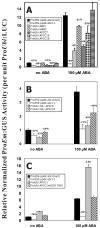
The 14,200 available full length Arabidopsis thaliana cDNAs in the universal plasmid system (UPS) donor vector pUNI51 should be applied broadly and efficiently to leverage a "functional map-space" of homologous plant genes. We have engineered Cre-lox UPS host acceptor vectors (pCR701- 705) with N-terminal epitope tags in frame with the loxH site and downstream from the maize Ubiquitin promoter for use in transient protoplast expression assays and particle bombardment transformation of monocots. As an example of the utility of these vectors, we recombined them with several Arabidopsis cDNAs encoding Ser/Thr protein phosphatase type 2C (PP2Cs) known from genetic studies or predicted by hierarchical clustering meta-analysis to be involved in ABA and stress responses. Our functional results in Zea mays mesophyll protoplasts on ABA-inducible expression effects on the Late Embryogenesis Abundant promoter ProEm:GUS reporter were consistent with predictions and resulted in identification of novel activities of some PP2Cs. Deployment of these vectors can facilitate functional genomics and proteomics and identification of novel gene activities.
Figures






Similar articles
-
Functional analyses of the ABI1-related protein phosphatase type 2C reveal evolutionarily conserved regulation of abscisic acid signaling between Arabidopsis and the moss Physcomitrella patens.Plant Mol Biol. 2009 Jun;70(3):327-40. doi: 10.1007/s11103-009-9476-z. Epub 2009 Mar 6. Plant Mol Biol. 2009. PMID: 19266168
-
Trivalent ions activate abscisic acid-inducible promoters through an ABI1-dependent pathway in rice protoplasts.Plant Physiol. 2000 Aug;123(4):1553-60. doi: 10.1104/pp.123.4.1553. Plant Physiol. 2000. PMID: 10938371 Free PMC article.
-
Arabidopsis ABA-Activated Kinase MAPKKK18 is Regulated by Protein Phosphatase 2C ABI1 and the Ubiquitin-Proteasome Pathway.Plant Cell Physiol. 2015 Dec;56(12):2351-67. doi: 10.1093/pcp/pcv146. Epub 2015 Oct 6. Plant Cell Physiol. 2015. PMID: 26443375 Free PMC article.
-
Signal transduction in maize and Arabidopsis mesophyll protoplasts.Plant Physiol. 2001 Dec;127(4):1466-75. Plant Physiol. 2001. PMID: 11743090 Free PMC article. Review.
-
Structural insights into PYR/PYL/RCAR ABA receptors and PP2Cs.Plant Sci. 2012 Jan;182:3-11. doi: 10.1016/j.plantsci.2010.11.014. Epub 2010 Dec 7. Plant Sci. 2012. PMID: 22118610 Review.
Cited by
-
Related to ABA-Insensitive3(ABI3)/Viviparous1 and AtABI5 transcription factor coexpression in cotton enhances drought stress adaptation.Plant Biotechnol J. 2014 Jun;12(5):578-89. doi: 10.1111/pbi.12162. Epub 2014 Feb 1. Plant Biotechnol J. 2014. PMID: 24483851 Free PMC article.
-
Identification and Candidate Gene Evaluation of a Large Fast Neutron-Induced Deletion Associated with a High-Oil Phenotype in Soybean Seeds.Genes (Basel). 2024 Jul 8;15(7):892. doi: 10.3390/genes15070892. Genes (Basel). 2024. PMID: 39062671 Free PMC article.
References
-
- Abremski K, Hoess R, Sternberg N. Studies on the properties of P1 site-specific recombination: evidence for topologically unlinked products following recombination. Cell. 1983;32:1301–1311. - PubMed
-
- Asselbergh B, De Vleesschauwer D, Hofte M. Global switches and fine-tuning - ABA modulates plant pathogen defense. Molec Plant-Microbe Int. 2008;21:709–719. - PubMed
-
- Ausubel FM. Summaries of National Science Foundation-sponsored Arabidopsis 2010 projects and National Science Foundation-sponsored plant genome projects that are generating Arabidopsis resources for the community. Plant Physiol. 2002;129:394–437.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

