AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading
- PMID: 19499576
- PMCID: PMC3041641
- DOI: 10.1002/jcc.21334
AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading
Abstract
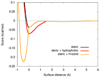
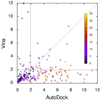
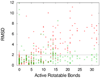
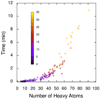
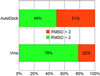
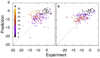
AutoDock Vina, a new program for molecular docking and virtual screening, is presented. AutoDock Vina achieves an approximately two orders of magnitude speed-up compared with the molecular docking software previously developed in our lab (AutoDock 4), while also significantly improving the accuracy of the binding mode predictions, judging by our tests on the training set used in AutoDock 4 development. Further speed-up is achieved from parallelism, by using multithreading on multicore machines. AutoDock Vina automatically calculates the grid maps and clusters the results in a way transparent to the user.
Copyright 2009 Wiley Periodicals, Inc.
Figures

References
-
- Sousa SF, Fernandes PA, Ramos MJ. Protein-ligand docking: Current status and future challenges. Proteins-Structure Function and Bioinformatics. 2006 Oct 1;vol. 65:15–26. - PubMed
-
- Chang MW, Lindstrom W, Olson AJ, Belew RK. Analysis of HIV wild-type and mutant structures via in silico docking against diverse ligand libraries. Journal of Chemical Information and Modeling. 2007 May–Jun;vol. 47:1258–1262. - PubMed
-
- Gilson MK, Zhou H-X. Calculation of protein-ligand binding affinities. Annual Review of Biophysics and Biomolecular Structure. 2007;vol. 36:21–42. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

