Epigenetic regulation in mammalian preimplantation embryo development
- PMID: 19500360
- PMCID: PMC2702308
- DOI: 10.1186/1477-7827-7-59
Epigenetic regulation in mammalian preimplantation embryo development
Abstract
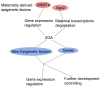
Preimplantation embryo development involves four stages: fertilization, cell cleavage, morula and blastocyst formation. During these stages, maternal and zygotic epigenetic factors play crucial roles. The gene expression profile is changed dramatically, chromatin is modified and core histone elements undergo significant changes. Each preimplantation embryo stage has its own characteristic epigenetic profile, consistent with the acquisition of the capacity to support development. Moreover, histone modifications such as methylation and acetylation as well as other epigenetic events can act as regulatory switches of gene transcription. Because the epigenetic profile is largely related to differentiation, epigenetic dysfunction can give rise to developmental abnormalities. Thus, epigenetic profiling of the embryo is of pivotal importance clinically. Given the importance of these aspects, this review will mainly focus on the epigenetic profile during preimplantation embryo development, as well as interactions between epigenetic and genetic regulation in these early developmental stages.
Figures

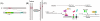

References
-
- Epstein CJ, Smith SA. Electrophoretic analysis of proteins synthesized by preimplantation mouse embryos. Dev Biol. 1974;40:233–244. - PubMed
-
- Weng DE, Morgan RA, Gearhart JD. Estimates of mRNA abundance in the mouse blastocyst based on cDNA library analysis. Mol Reprod Dev. 1989;1:233–241. - PubMed
-
- Ma J, Svoboda P, Schultz RM, Stein P. Regulation of Zygotic Gene Activation in the Preimplantation Mouse Embryo: Global Activation and Repression of Gene Expression. Biol Reprod. 2001;64:1713–1721. - PubMed
-
- Zeng F, Schultz RM. Gene expression in mouse oocytes and preimplantation embryos: use of suppression subtractive hybridization to identify oocyte- and embryo-specific genes. Biol Reprod. 2003;68:31–39. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

