Biochemical evidence for a timing mechanism in prochlorococcus
- PMID: 19502405
- PMCID: PMC2725622
- DOI: 10.1128/JB.00419-09
Biochemical evidence for a timing mechanism in prochlorococcus
Abstract
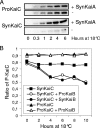
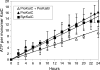
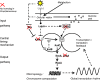
Organisms coordinate biological activities into daily cycles using an internal circadian clock. The circadian oscillator proteins KaiA, KaiB, and KaiC are widely believed to underlie 24-h oscillations of gene expression in cyanobacteria. However, a group of very abundant cyanobacteria, namely, marine Prochlorococcus species, lost the third oscillator component, KaiA, during evolution. We demonstrate here that the remaining Kai proteins fulfill their known biochemical functions, although KaiC is hyperphosphorylated by default in this system. These data provide biochemical support for the observed evolutionary reduction of the clock locus in Prochlorococcus and are consistent with a model in which a mechanism that is less robust than the well-characterized KaiABC protein clock of Synechococcus is sufficient for biological timing in the very stable environment that Prochlorococcus inhabits.
Figures
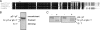
Comment in
-
The rolex and the hourglass: a simplified circadian clock in prochlorococcus?J Bacteriol. 2009 Sep;191(17):5333-5. doi: 10.1128/JB.00719-09. Epub 2009 Jun 26. J Bacteriol. 2009. PMID: 19561127 Free PMC article. No abstract available.
References
-
- Dufresne, A., M. Salanoubat, F. Partensky, F. Artiguenave, I. M. Axmann, V. Barbe, S. Duprat, M. Y. Galperin, E. V. Koonin, F. Le Gall, K. S. Makarova, M. Ostrowski, S. Oztas, C. Robert, I. B. Rogozin, D. J. Scanlan, N. Tandeau de Marsac, J. Weissenbach, P. Wincker, Y. I. Wolf, and W. R. Hess. 2003. Genome sequence of the cyanobacterium Prochlorococcus marinus SS120, a nearly minimal oxyphototrophic genome. Proc. Natl. Acad. Sci. USA 10010020-10025. - PMC - PubMed
-
- Goericke, R., and N. A. Welschmeyer. 1993. The marine prochlorophyte Prochlorococcus contributes significantly to phytoplankton biomass and primary production in the Sargasso Sea. Deep-Sea Res. 402283-2294.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

