Activation of alternative Jdp2 promoters and functional protein isoforms in T-cell lymphomas by retroviral insertion mutagenesis
- PMID: 19502497
- PMCID: PMC2724284
- DOI: 10.1093/nar/gkp469
Activation of alternative Jdp2 promoters and functional protein isoforms in T-cell lymphomas by retroviral insertion mutagenesis
Abstract
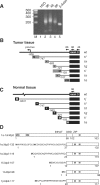
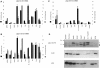
Retroviral insertional mutagenesis has been instrumental for the identification of genes important in cancer development. The molecular mechanisms involved in retroviral-mediated activation of proto-oncogenes influence the distribution of insertions within specific regions during tumorigenesis and hence may point to novel gene structures. From a retroviral tagging screen on tumors of 1767 SL3-3 MLV-infected BALB/c mice, intron 2 of the AP-1 repressor Jdp2 locus was found frequently targeted by proviruses resulting in upregulation of non-canonical RNA subspecies. We identified several promoter regions within 1000 bp upstream of exon 3 that allowed for the production of Jdp2 protein isoforms lacking the histone acetylase inhibitory domain INHAT present in canonical Jdp2. The novel Jdp2 isoforms localized to the nucleus and over-expression in murine fibroblast cells induced cell death similar to canonic Jdp2. When expressed in the context of oncogenic NRAS both full length Jdp2 and the shorter isoforms increased anchorage-independent growth. Our results demonstrate a biological function of Jdp2 lacking the INHAT domain and suggest a post-genomic application for the use of retroviral tagging data in identifying new gene products with a potential role in tumorigenesis.
Figures





References
-
- Jin C, Ugai H, Song J, Murata T, Nili F, Sun K, Horikoshi M, Yokoyama KK. Identification of mouse Jun dimerization protein 2 as a novel repressor of ATF-2. FEBS Lett. 2001;489:34–41. - PubMed
-
- Broder YC, Katz S, Aronheim A. The ras recruitment system, a novel approach to the study of protein–protein interactions. Curr. Biol. 1998;8:1121–1124. - PubMed
-
- Cherasse Y, Chaveroux C, Jousse C, Maurin AC, Carraro V, Parry L, Fafournoux P, Bruhat A. Role of the repressor JDP2 in the amino acid-regulated transcription of CHOP. FEBS Lett. 2008;582:1537–1541. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

