Structure of complement fragment C3b-factor H and implications for host protection by complement regulators
- PMID: 19503104
- PMCID: PMC2713992
- DOI: 10.1038/ni.1755
Structure of complement fragment C3b-factor H and implications for host protection by complement regulators
Abstract
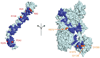
Factor H (FH) is an abundant regulator of complement activation and protects host cells from self-attack by complement. Here we provide insight into the regulatory activity of FH by solving the crystal structure of the first four domains of FH in complex with its target, complement fragment C3b. FH interacted with multiple domains of C3b, covering a large, extended surface area. The structure indicated that FH destabilizes the C3 convertase by competition and electrostatic repulsion and that FH enables proteolytic degradation of C3b by providing a binding platform for protease factor I while stabilizing the overall domain arrangement of C3b. Our results offer general models for complement regulation and provide structural explanations for disease-related mutations in the genes encoding both FH and C3b.
Figures

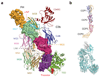


References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

