Pervasive natural selection in the Drosophila genome?
- PMID: 19503600
- PMCID: PMC2684638
- DOI: 10.1371/journal.pgen.1000495
Pervasive natural selection in the Drosophila genome?
Abstract
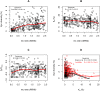
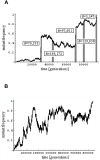
Over the past four decades, the predominant view of molecular evolution saw little connection between natural selection and genome evolution, assuming that the functionally constrained fraction of the genome is relatively small and that adaptation is sufficiently infrequent to play little role in shaping patterns of variation within and even between species. Recent evidence from Drosophila, reviewed here, suggests that this view may be invalid. Analyses of genetic variation within and between species reveal that much of the Drosophila genome is under purifying selection, and thus of functional importance, and that a large fraction of coding and noncoding differences between species are adaptive. The findings further indicate that, in Drosophila, adaptations may be both common and strong enough that the fate of neutral mutations depends on their chance linkage to adaptive mutations as much as on the vagaries of genetic drift. The emerging evidence has implications for a wide variety of fields, from conservation genetics to bioinformatics, and presents challenges to modelers and experimentalists alike.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

