The role of geography in human adaptation
- PMID: 19503611
- PMCID: PMC2685456
- DOI: 10.1371/journal.pgen.1000500
The role of geography in human adaptation
Abstract
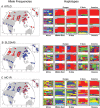
Various observations argue for a role of adaptation in recent human evolution, including results from genome-wide studies and analyses of selection signals at candidate genes. Here, we use genome-wide SNP data from the HapMap and CEPH-Human Genome Diversity Panel samples to study the geographic distributions of putatively selected alleles at a range of geographic scales. We find that the average allele frequency divergence is highly predictive of the most extreme F(ST) values across the whole genome. On a broad scale, the geographic distribution of putatively selected alleles almost invariably conforms to population clusters identified using randomly chosen genetic markers. Given this structure, there are surprisingly few fixed or nearly fixed differences between human populations. Among the nearly fixed differences that do exist, nearly all are due to fixation events that occurred outside of Africa, and most appear in East Asia. These patterns suggest that selection is often weak enough that neutral processes -- especially population history, migration, and drift -- exert powerful influences over the fate and geographic distribution of selected alleles.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
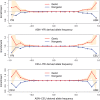
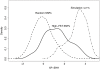
 in derived allele frequency between two HapMap populations. The y-axis plots the fold enrichment of genic and nongenic SNPs as a function of
in derived allele frequency between two HapMap populations. The y-axis plots the fold enrichment of genic and nongenic SNPs as a function of  : i.e., for each bin we plot the fraction of SNPs in that bin that are genic (respectively, nongenic), divided by the fraction of all SNPs that are genic (respectively, nongenic). The peach-colored region gives the central 90% confidence interval (estimated by bootstrap resampling of 200 kb regions from the genome); when the lower edge of the peach region is >1 this indicates significant enrichment of genic SNPs, assuming a one-tailed test at p = 0.05. Genotype frequencies were estimated from Phase II HapMap data using only SNPs that were identified by Perlegen in a uniform multiethnic panel (“Type A” SNPs) . The numbers of SNPs in the tails are given in Supplementary Table 1 in Text S1.
: i.e., for each bin we plot the fraction of SNPs in that bin that are genic (respectively, nongenic), divided by the fraction of all SNPs that are genic (respectively, nongenic). The peach-colored region gives the central 90% confidence interval (estimated by bootstrap resampling of 200 kb regions from the genome); when the lower edge of the peach region is >1 this indicates significant enrichment of genic SNPs, assuming a one-tailed test at p = 0.05. Genotype frequencies were estimated from Phase II HapMap data using only SNPs that were identified by Perlegen in a uniform multiethnic panel (“Type A” SNPs) . The numbers of SNPs in the tails are given in Supplementary Table 1 in Text S1.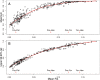
 ) for each population pair, and (B) the value of the 65th most extreme
) for each population pair, and (B) the value of the 65th most extreme  for each population pair (i.e., the 99.99th percentile of the allele frequency distribution). To provide a sense of scale on the figure, red arrows are used to indicate the mean autosomal pairwise FST between some arbitrary pairs of populations (key: French (Fra), Palestinian (Pal), Han-Chinese (Han) and Yoruba (Yor)). The red lines plot lowess fits to the data. Plots of the extremes of pairwise FST and with different sample size cutoffs are similar (Supplementary Figures 5 and 6 in Text S1).
for each population pair (i.e., the 99.99th percentile of the allele frequency distribution). To provide a sense of scale on the figure, red arrows are used to indicate the mean autosomal pairwise FST between some arbitrary pairs of populations (key: French (Fra), Palestinian (Pal), Han-Chinese (Han) and Yoruba (Yor)). The red lines plot lowess fits to the data. Plots of the extremes of pairwise FST and with different sample size cutoffs are similar (Supplementary Figures 5 and 6 in Text S1).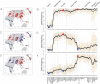
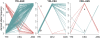

 at time 0, conditional on the alleles not being lost within 4000 generations. Simulations were performed under an effective population size of 24,000 chosen to match the effective population size of the ‘Yoruba’ in cosi
. To provide some context, the bars at the top indicate the divergence times of the HapMap Europeans and Asians, and HapMap Africans and non-Africans according to the cosi model , though it should be noted that there is considerable uncertainty in the true split times. The numbers in parentheses indicate times in years, assuming 20 years per generation.
at time 0, conditional on the alleles not being lost within 4000 generations. Simulations were performed under an effective population size of 24,000 chosen to match the effective population size of the ‘Yoruba’ in cosi
. To provide some context, the bars at the top indicate the divergence times of the HapMap Europeans and Asians, and HapMap Africans and non-Africans according to the cosi model , though it should be noted that there is considerable uncertainty in the true split times. The numbers in parentheses indicate times in years, assuming 20 years per generation.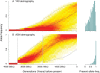
References
-
- Sabeti P, Schaffner S, Fry B, Lohmueller J, Varilly P, et al. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. - PubMed
-
- Volkman S, Sabeti P, DeCaprio D, Neafsey D, Schaffner S, et al. A genome-wide map of diversity in Plasmodium falciparum. Nat Genet. 2007;39:113–119. - PubMed
-
- Clark R, Schweikert G, Toomajian C, Ossowski S, Zeller G, et al. Common sequence poly-morphisms shaping genetic diversity in Arabidopsis thaliana. Science. 2007;317:338–342. - PubMed
-
- Stringer C, Andrews P. Genetic and fossil evidence for the origin of modern humans. Science. 1988;239:1263–1268. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

