Failure to replicate a genetic association may provide important clues about genetic architecture
- PMID: 19503614
- PMCID: PMC2685469
- DOI: 10.1371/journal.pone.0005639
Failure to replicate a genetic association may provide important clues about genetic architecture
Abstract
Replication has become the gold standard for assessing statistical results from genome-wide association studies. Unfortunately this replication requirement may cause real genetic effects to be missed. A real result can fail to replicate for numerous reasons including inadequate sample size or variability in phenotype definitions across independent samples. In genome-wide association studies the allele frequencies of polymorphisms may differ due to sampling error or population differences. We hypothesize that some statistically significant independent genetic effects may fail to replicate in an independent dataset when allele frequencies differ and the functional polymorphism interacts with one or more other functional polymorphisms. To test this hypothesis, we designed a simulation study in which case-control status was determined by two interacting polymorphisms with heritabilities ranging from 0.025 to 0.4 with replication sample sizes ranging from 400 to 1600 individuals. We show that the power to replicate the statistically significant independent main effect of one polymorphism can drop dramatically with a change of allele frequency of less than 0.1 at a second interacting polymorphism. We also show that differences in allele frequency can result in a reversal of allelic effects where a protective allele becomes a risk factor in replication studies. These results suggest that failure to replicate an independent genetic effect may provide important clues about the complexity of the underlying genetic architecture. We recommend that polymorphisms that fail to replicate be checked for interactions with other polymorphisms, particularly when samples are collected from groups with distinct ethnic backgrounds or different geographic regions.
Conflict of interest statement
Figures

 ) is enough to drop the power to replicate
) is enough to drop the power to replicate  from 80% to 20% for this model. It is apparent in B that as the minor allele frequency,
from 80% to 20% for this model. It is apparent in B that as the minor allele frequency,  , of
, of  in the sampled population moves from 0.3 to 0.5 the marginal penetrances of the alleles for
in the sampled population moves from 0.3 to 0.5 the marginal penetrances of the alleles for  (
( ) become equal and the main effect is lost. When the replication sample is performed at an allele frequency of 0.3 the power to detect a main effect is near 100%, at an allele frequency of 0.4 the power to detect a main effect is near 60%, and at an allele frequency of 0.5 the marginal penetrances are equivalent and no main effect remains.
) become equal and the main effect is lost. When the replication sample is performed at an allele frequency of 0.3 the power to detect a main effect is near 100%, at an allele frequency of 0.4 the power to detect a main effect is near 60%, and at an allele frequency of 0.5 the marginal penetrances are equivalent and no main effect remains.
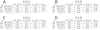
References
-
- Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6:95–108. - PubMed
-
- Wang WYS, Barratt BJ, Clayton DG, Todd JA. Genome-wide association studies: theoretical and practical concerns. Nat Rev Genet. 2005;6:109–118. - PubMed
-
- Pe'er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genetic Epidemiology. 2008;32:381–385. - PubMed
-
- Chanock SJ, Manolio T, Boehnke M, Boerwinkle E, Hunter DJ, et al. Replicating genotypephenotype associations. Nature. 2007;447:655–60. - PubMed
-
- Shriner D, Vaughan LK, Padilla MA, Tiwari HK. Problems with Genome-Wide association studies. Science. 2007;316:1840–1841. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

