Fgf and Sdf-1 pathways interact during zebrafish fin regeneration
- PMID: 19503807
- PMCID: PMC2688747
- DOI: 10.1371/journal.pone.0005824
Fgf and Sdf-1 pathways interact during zebrafish fin regeneration
Abstract
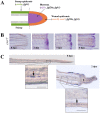
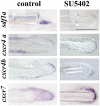
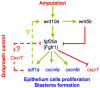
The chemokine stromal cell-derived factor-1 (SDF1) was originally identified as a pre-B cell stimulatory factor but has been recently implicated in several other key steps in differentiation and morphogenesis. In addition, SDF1 as well as FGF signalling pathways have recently been shown to be involved in the control of epimorphic regeneration. In this report, we address the question of a possible interaction between the two signalling pathways during adult fin regeneration in zebrafish. Using a combination of pharmaceutical and genetic tools, we show that during epimorphic regeneration, expression of sdf1, as well as of its cognate receptors, cxcr4a, cxcr4b and cxcr7 are controlled by FGF signalling. We further show that, Sdf1a negatively regulates the expression of fgf20a. Together, these results lead us to propose that: 1) the function of Fgf in blastema formation is, at least in part, relayed by the chemokine Sdf1a, and that 2) Sdf1 exerts negative feedback on the Fgf pathway, which contributes to a transient expression of Fgf20a downstream genes at the beginning of regeneration. However this feedback control can be bypassed since the Sdf1 null mutants regenerate their fin, though slower. Very few mutants for the regeneration process were isolated so far, illustrating the difficulty in identifying genes that are indispensable for regeneration. This observation supports the idea that the regeneration process involves a delicate balance between multiple pathways.
Conflict of interest statement
Figures

References
-
- Nakatani Y, Kawakami A, Kudo A. Cellular and molecular processes of regeneration, with special emphasis on fish fins. Dev Growth Differ. 2007;49:145–154. - PubMed
-
- Poss KD, Keating MT, Nechiporuk A. Tales of regeneration in zebrafish. Dev Dyn. 2003;226:202–210. - PubMed
-
- Akimenko MA, Mari-Beffa M, Becerra J, Geraudie J. Old questions, new tools, and some answers to the mystery of fin regeneration. Dev Dyn. 2003;226:190–201. - PubMed
-
- Slack JM, Tannahill D. Mechanism of anteroposterior axis specification in vertebrates. Lessons from the amphibians. Development. 1992;114:285–302. - PubMed
-
- Slack J. Role of fibroblast growth factors as inducing agents in early embryonic development. Mol Reprod Dev. 1994;39:118–124. discussion 124–115. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

