The two active sites of Thermotoga maritima CheA dimers bind ATP with dramatically different affinities
- PMID: 19505148
- PMCID: PMC2811258
- DOI: 10.1021/bi900474g
The two active sites of Thermotoga maritima CheA dimers bind ATP with dramatically different affinities
Abstract
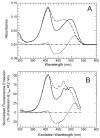
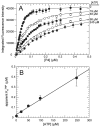
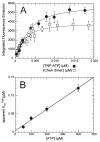
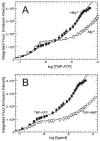
CheA is a central component of the chemotaxis signal transduction pathway that allows prokaryotic cells to control their movements in response to environmental cues. This dimeric protein histidine kinase autophosphorylates via an intersubunit phosphorylation reaction in which each protomer of the dimer binds ATP, at an active site located in its P4 domain and then catalyzes transfer of the gamma-phosphoryl group of ATP to the His(45) side chain within the P1 domain of the trans protomer. Here we utilize the fluorescent nucleotide analogue TNP-ATP [2'(3')-O-(2,4,6-trinitrophenyl)adenosine 5'-triphosphate] to investigate the two ATP-binding sites of the Thermotoga maritima CheA dimer (TmCheA) and the single site of the isolated TmP4 domain (a monomer). We define the affinity of CheA for TNP nucleotides and, by competition, for unmodified ATP. The two ATP-binding sites of the TmCheA dimer exhibit dramatically different affinities for TNP-ATP (K(d1)(TNP) approximately 0.0016 muM and K(d2)(TNP) approximately 22 muM at 4 degrees C in the presence of Mg(2+)) as well as for ATP (K(d1)(ATP) approximately 6 muM and K(d2)(ATP) approximately 5000 muM at 4 degrees C in the presence of Mg(2+)) and in their ability to influence the fluorescence of bound TNP-ATP. The ATP-binding site of the isolated TmP4 domain interacts with ATP and TNP-ATP in a manner similar to that of the high-affinity site of the TmCheA dimer. These results suggest that the two active sites of TmCheA homodimers exhibit large differences in their interactions with ATP. We consider possible implications of these differences for the CheA autophosphorylation mechanism and for CheA function in bacterial cells.
Figures




Similar articles
-
TNP-ATP and TNP-ADP as probes of the nucleotide binding site of CheA, the histidine protein kinase in the chemotaxis signal transduction pathway of Escherichia coli.Biochemistry. 1998 Sep 1;37(35):12269-79. doi: 10.1021/bi980970n. Biochemistry. 1998. PMID: 9724541
-
Kinetics of ATP and TNP-ATP binding to the active site of CheA from Thermotoga maritima.Biochemistry. 2010 Jul 13;49(27):5799-809. doi: 10.1021/bi100721b. Biochemistry. 2010. PMID: 20565117
-
Conformational Transitions that Enable Histidine Kinase Autophosphorylation and Receptor Array Integration.J Mol Biol. 2015 Dec 4;427(24):3890-907. doi: 10.1016/j.jmb.2015.10.015. Epub 2015 Oct 30. J Mol Biol. 2015. PMID: 26522934 Free PMC article.
-
Nucleotide binding by the histidine kinase CheA.Nat Struct Biol. 2001 Apr;8(4):353-60. doi: 10.1038/86243. Nat Struct Biol. 2001. PMID: 11276258
-
Histidine kinases: diversity of domain organization.Mol Microbiol. 1999 Nov;34(4):633-40. doi: 10.1046/j.1365-2958.1999.01646.x. Mol Microbiol. 1999. PMID: 10564504 Review.
Cited by
-
Engineered chemotaxis core signaling units indicate a constrained kinase-off state.Sci Signal. 2020 Nov 10;13(657):eabc1328. doi: 10.1126/scisignal.abc1328. Sci Signal. 2020. PMID: 33172954 Free PMC article.
-
Cell fate regulation governed by a repurposed bacterial histidine kinase.PLoS Biol. 2014 Oct 28;12(10):e1001979. doi: 10.1371/journal.pbio.1001979. eCollection 2014 Oct. PLoS Biol. 2014. PMID: 25349992 Free PMC article.
-
Nucleotide Spin Labeling for ESR Spectroscopy of ATP-Binding Proteins.Appl Magn Reson. 2018 Dec;49(12):1385-1395. doi: 10.1007/s00723-018-1070-6. Epub 2018 Nov 14. Appl Magn Reson. 2018. PMID: 30686862 Free PMC article.
-
Substrate-modulated thermal fluctuations affect long-range allosteric signaling in protein homodimers: exemplified in CAP.Biophys J. 2010 May 19;98(10):2317-26. doi: 10.1016/j.bpj.2010.01.039. Biophys J. 2010. PMID: 20483341 Free PMC article.
-
Structure of the ternary complex formed by a chemotaxis receptor signaling domain, the CheA histidine kinase, and the coupling protein CheW as determined by pulsed dipolar ESR spectroscopy.Biochemistry. 2010 May 11;49(18):3824-41. doi: 10.1021/bi100055m. Biochemistry. 2010. PMID: 20355710 Free PMC article.
References
-
- Bilwes A, Alex L, Crane BR, Simon MI. Structure of CheA, a signal-transducing histidine kinase. Cell. 1999;96:131–141. - PubMed
-
- Bilwes A, Quezada CM, Croal LR, Crane BR, Simon MI. Nucleotide binding by the histidine kinase CheA. Nat Struct Biol. 2001;8:353–360. - PubMed
-
- Bilwes AMP, Quezada CM, Simon M, Crane BR. Histidine Kinases in Signal Transduction. Academic Press; San Diego: 2003. pp. 48–74.
-
- Borkovich KA, Simon MI. The dynamics of protein phosphorylation in bacterial chemotaxis. Cell. 1990;63:1339–1348. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

