A clustering approach for identification of enriched domains from histone modification ChIP-Seq data
- PMID: 19505939
- PMCID: PMC2732366
- DOI: 10.1093/bioinformatics/btp340
A clustering approach for identification of enriched domains from histone modification ChIP-Seq data
Abstract
Motivation: Chromatin states are the key to gene regulation and cell identity. Chromatin immunoprecipitation (ChIP) coupled with high-throughput sequencing (ChIP-Seq) is increasingly being used to map epigenetic states across genomes of diverse species. Chromatin modification profiles are frequently noisy and diffuse, spanning regions ranging from several nucleosomes to large domains of multiple genes. Much of the early work on the identification of ChIP-enriched regions for ChIP-Seq data has focused on identifying localized regions, such as transcription factor binding sites. Bioinformatic tools to identify diffuse domains of ChIP-enriched regions have been lacking.
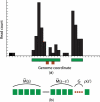
Results: Based on the biological observation that histone modifications tend to cluster to form domains, we present a method that identifies spatial clusters of signals unlikely to appear by chance. This method pools together enrichment information from neighboring nucleosomes to increase sensitivity and specificity. By using genomic-scale analysis, as well as the examination of loci with validated epigenetic states, we demonstrate that this method outperforms existing methods in the identification of ChIP-enriched signals for histone modification profiles. We demonstrate the application of this unbiased method in important issues in ChIP-Seq data analysis, such as data normalization for quantitative comparison of levels of epigenetic modifications across cell types and growth conditions.
Availability: http://home.gwu.edu/ approximately wpeng/Software.htm.
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures



References
-
- Bannister AJ, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. - PubMed
-
- Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995;57:289–300.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

