Evolution and diversity of glutaredoxins in photosynthetic organisms
- PMID: 19506802
- PMCID: PMC11115520
- DOI: 10.1007/s00018-009-0054-y
Evolution and diversity of glutaredoxins in photosynthetic organisms
Abstract
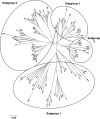
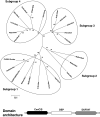
The genome sequencing of prokaryotic and eukaryotic photosynthetic organisms enables a comparative genomic study of the glutaredoxin (Grx) family. The analysis of 58 genomes, using a specific motif composed of the active site sequence and of amino acids involved in glutathione binding, led to an updated classification of Grxs into six classes. Only two classes (I and II) are common to all photosynthetic organisms. Eukaryotes and cyanobacteria have two specific Grx classes (classes III and IV and classes V and VI, respectively). The classes IV, V and VI have not yet been identified and contain multimodular Grx fusions. In addition, putative Grx partners were identified from the presence of fusion proteins, the conservation of gene order in bacterial operons, and the gene co-occurrence. The genes encoding class II Grxs and BolA/YrbA proteins are frequently adjacent, in the same transcriptional orientation in prokaryote genomes and present in the same organisms.
Figures







Similar articles
-
Phylogenetic distribution and structural analyses of cyanobacterial glutaredoxins (Grxs).Comput Biol Chem. 2020 Feb;84:107141. doi: 10.1016/j.compbiolchem.2019.107141. Epub 2019 Dec 12. Comput Biol Chem. 2020. PMID: 31839562
-
Biochemical characterization of glutaredoxins from Chlamydomonas reinhardtii reveals the unique properties of a chloroplastic CGFS-type glutaredoxin.J Biol Chem. 2008 Apr 4;283(14):8868-76. doi: 10.1074/jbc.M709567200. Epub 2008 Jan 23. J Biol Chem. 2008. PMID: 18216016
-
Evolution based on domain combinations: the case of glutaredoxins.BMC Evol Biol. 2009 Mar 25;9:66. doi: 10.1186/1471-2148-9-66. BMC Evol Biol. 2009. PMID: 19321008 Free PMC article.
-
The biological roles of glutaredoxins.Biochem J. 2012 Sep 15;446(3):333-48. doi: 10.1042/BJ20112131. Biochem J. 2012. PMID: 22928493 Review.
-
Glutaredoxins with iron-sulphur clusters in eukaryotes - Structure, function and impact on disease.Biochim Biophys Acta Bioenerg. 2021 Jan 1;1862(1):148317. doi: 10.1016/j.bbabio.2020.148317. Epub 2020 Sep 24. Biochim Biophys Acta Bioenerg. 2021. PMID: 32980338 Review.
Cited by
-
BOLA1 is an aerobic protein that prevents mitochondrial morphology changes induced by glutathione depletion.Antioxid Redox Signal. 2013 Jan 10;18(2):129-38. doi: 10.1089/ars.2011.4253. Epub 2012 Sep 11. Antioxid Redox Signal. 2013. PMID: 22746225 Free PMC article.
-
Deciphering the mechanism of glutaredoxin-catalyzed roGFP2 redox sensing reveals a ternary complex with glutathione for protein disulfide reduction.Nat Commun. 2024 Feb 26;15(1):1733. doi: 10.1038/s41467-024-45808-9. Nat Commun. 2024. PMID: 38409212 Free PMC article.
-
Glutaredoxin S15 Is Involved in Fe-S Cluster Transfer in Mitochondria Influencing Lipoic Acid-Dependent Enzymes, Plant Growth, and Arsenic Tolerance in Arabidopsis.Plant Physiol. 2016 Mar;170(3):1284-99. doi: 10.1104/pp.15.01308. Epub 2015 Dec 15. Plant Physiol. 2016. PMID: 26672074 Free PMC article.
-
Glutaredoxin: Discovery, redox defense and much more.Redox Biol. 2021 Jul;43:101975. doi: 10.1016/j.redox.2021.101975. Epub 2021 Apr 20. Redox Biol. 2021. PMID: 33932870 Free PMC article. Review.
-
Protein-thiol oxidation and cell death: regulatory role of glutaredoxins.Antioxid Redox Signal. 2012 Dec 15;17(12):1748-63. doi: 10.1089/ars.2012.4644. Epub 2012 Jun 5. Antioxid Redox Signal. 2012. PMID: 22530666 Free PMC article. Review.
References
-
- Fernandes AP, Fladvad M, Berndt C, Andresen C, Lillig CH, Neubauer P, Sunnerhagen M, Holmgren A, Vlamis-Gardikas A. A novel monothiol glutaredoxin (Grx4) from Escherichia coli can serve as a substrate for thioredoxin reductase. J Biol Chem. 2005;280:24544–24552. doi: 10.1074/jbc.M500678200. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

