eIF2alpha kinases GCN2 and PERK modulate transcription and translation of distinct sets of mRNAs in mouse liver
- PMID: 19509078
- PMCID: PMC3171828
- DOI: 10.1152/physiolgenomics.90396.2008
eIF2alpha kinases GCN2 and PERK modulate transcription and translation of distinct sets of mRNAs in mouse liver
Abstract
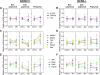
In eukaryotes, selective derepression of mRNA translation through altered utilization of upstream open reading frames (uORF) or internal ribosomal entry sites (IRES) regulatory motifs following exposure to stress is regulated at the initiation stage through the increased phosphorylation of eukaryotic initiation factor 2 on its alpha-subunit (eIF2alpha). While there is only one known eIF2alpha kinase in yeast, general control nonderepressible 2 (GCN2), mammals have evolved to express at least four: GCN2, heme-regulated inhibitor kinase (HRI), double-stranded RNA-activated protein kinase (PKR), and PKR-like endoplasmic reticulum-resident kinase (PERK). So far, the main known distinction among these four kinases is their activation in response to different acute stressors. In the present study, we used the in situ perfused mouse liver model and hybridization array analyses to assess the general translational response to stress regulated by two of these kinases, GCN2 and PERK, and to differentiate between the downstream effects of activating GCN2 versus PERK. The resulting data showed that at least 2.5% of mouse liver mRNAs are subject to derepressed translation following stress. In addition, the data demonstrated that eIF2alpha kinases GCN2 and PERK differentially regulate mRNA transcription and translation, which in the latter case suggests that increased eIF2alpha phosphorylation is not sufficient for derepression of translation. These findings open an avenue for more focused future research toward groups of mRNAs that code for the early cellular stress response proteins.
Figures
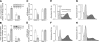
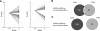






References
-
- Al Sarraj J, Vinson C, Thiel G. Regulation of asparagine synthetase gene transcription by the basic region leucine zipper transcription factors ATF5 and CHOP. J Biol Chem 386: 873–879, 2005. - PubMed
-
- Anthony TG, Reiter AK, Anthony JC, Kimball SR, Jefferson LS. Deficiency of dietary EAA preferentially inhibits mRNA translation of ribosomal proteins in liver of meal-fed rats. Am J Physiol Endocrinol Metab 281: E430–E439, 2001. - PubMed
-
- Averous J, Maurin AC, Bruhat A, Jousse C, Arliguie C, Fafournoux P. Induction of IGFBP-1 expression by amino acid deprivation of HepG2 human hepatoma cells involves both a transcriptional activation and an mRNA stabilization due to its 3′UTR. FEBS Lett 579: 2609–2614, 2005. - PubMed
-
- Baltzis D, Pluquet O, Papadakis AI, Kazemi S, Qu LK, Koromilas AE. The eIF2α kinases PERK and PKR activate glycogen synthase kinase 3 to promote the proteasomal degradation of p53. J Biol Chem 282: 31675–31687, 2007. - PubMed
-
- Bi M, Naczki C, Koritzinsky M, Fels D, Blais J, Hu N, Harding H, Novoa I, Varia M, Raleigh J, Scheuner D, Kaufman RJ, Bell J, Ron D, Wouters BG, Koumenis C, Bi M, Naczki C, Koritzinsky M, Fels D, Blais J, Hu N, Harding H, Novoa I, Varia M, Raleigh J, Scheuner D, Kaufman RJ, Bell J, Ron D, Wouters BG, Koumenis C. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J 24: 3470–3481, 2005. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

