Modulation of TLR2 protein expression by miR-105 in human oral keratinocytes
- PMID: 19509287
- PMCID: PMC2755716
- DOI: 10.1074/jbc.M109.013862
Modulation of TLR2 protein expression by miR-105 in human oral keratinocytes
Abstract
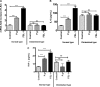
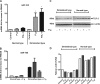
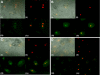
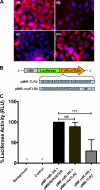
Mammalian biological processes such as inflammation, involve regulation of hundreds of genes controlling onset and termination. MicroRNAs (miRNAs) can translationally repress target mRNAs and regulate innate immune responses. Our model system comprised primary human keratinocytes, which exhibited robust differences in inflammatory cytokine production (interleukin-6 and tumor necrosis factor-alpha) following specific Toll-like receptor 2 and 4 (TLR-2/TLR-4) agonist challenge. We challenged these primary cells with Porphyromonas gingivalis (a Gram-negative bacterium that triggers TLR-2 and TLR-4) and performed miRNA expression profiling. We identified miRNA (miR)-105 as a modulator of TLR-2 protein translation in human gingival keratinocytes. There was a strong inverse correlation between cells that had high cytokine responses following TLR-2 agonist challenge and miR-105 levels. Knock-in and knock-down of miR-105 confirmed this inverse relationship. In silico analysis predicted that miR-105 had complementarity for TLR-2 mRNA, and the luciferase reporter assay verified this. Further understanding of the role of miRNA in host responses may elucidate disease susceptibility and suggest new anti-inflammatory therapeutics.
Figures


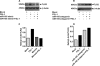
References
-
- Akira S., Uematsu S., Takeuchi O. (2006) Cell 124,783–801 - PubMed
-
- Kinane D. F., Shiba H., Stathopoulou P. G., Zhao H., Lappin D. F., Singh A., Eskan M. A., Beckers S., Waigel S., Alpert B., Knudsen T. B. (2006) Genes Immun. 7,190–200 - PubMed
-
- Akira S., Takeda K. (2004) Nat. Rev. Immunol. 4,499–511 - PubMed
-
- Bartel D. P. (2004) Cell 116,281–297 - PubMed
-
- Liew F. Y., Xu D., Brint E. K., O'Neill L. A. (2005) Nat. Rev. Immunol. 5,446–458 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

