Kinetic and functional analysis of L-threonine kinase, the PduX enzyme of Salmonella enterica
- PMID: 19509296
- PMCID: PMC2740450
- DOI: 10.1074/jbc.M109.027425
Kinetic and functional analysis of L-threonine kinase, the PduX enzyme of Salmonella enterica
Abstract
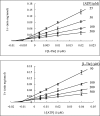
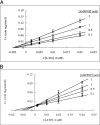
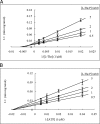
The PduX enzyme of Salmonella enterica is an l-threonine kinase used for the de novo synthesis of coenzyme B(12) and the assimilation of cobyric acid. PduX with an N-terminal histidine tag (His(8)-PduX) was produced in Escherichiacoli and purified. The recombinant enzyme was soluble and active. Kinetic analysis indicated a steady-state Ordered Bi Bi complex mechanism in which ATP is the first substrate to bind. Based on a multiple sequence alignment of PduX homologues and other GHMP (galactokinase, homoserine kinase, mevalonate kinase, and phosphomevalonate kinase) family members, 14 PduX variants having changes at 10 conserved serine/threonine and aspartate/glutamate sites were constructed by site-directed mutagenesis. Each variant was produced in E. coli and purified. Comparison of the circular dichroism spectra and kinetic properties of the PduX variants with those of the wild-type enzyme indicated that Glu-24 and Asp-135 are needed for proper folding, Ser-99 and Glu-132 are used for ATP binding, and Ser-253 and Ser-255 are critical to l-threonine binding whereas Ser-100 is essential to catalysis, but its precise role is uncertain. The studies reported here are the first to investigate the kinetic and catalytic mechanisms of l-threonine kinase from any organism.
Figures






Similar articles
-
The PduX enzyme of Salmonella enterica is an L-threonine kinase used for coenzyme B12 synthesis.J Biol Chem. 2008 Apr 25;283(17):11322-9. doi: 10.1074/jbc.M800287200. Epub 2008 Feb 28. J Biol Chem. 2008. PMID: 18308727
-
Investigation of invariant serine/threonine residues in mevalonate kinase. Tests of the functional significance of a proposed substrate binding motif and a site implicated in human inherited disease.J Biol Chem. 2001 Apr 20;276(16):12573-8. doi: 10.1074/jbc.M011478200. Epub 2001 Jan 17. J Biol Chem. 2001. PMID: 11278915
-
Mass spectrometry and site-directed mutagenesis identify several autophosphorylated residues required for the activity of PrkC, a Ser/Thr kinase from Bacillus subtilis.J Mol Biol. 2003 Jul 11;330(3):459-72. doi: 10.1016/s0022-2836(03)00579-5. J Mol Biol. 2003. PMID: 12842463
-
Rhodobacterales use a unique L-threonine kinase for the assembly of the nucleotide loop of coenzyme B12.Mol Microbiol. 2018 Oct;110(2):239-261. doi: 10.1111/mmi.14100. Epub 2018 Oct 3. Mol Microbiol. 2018. PMID: 30098062 Free PMC article.
-
Investigation of a catalytic zinc binding site in Escherichia coli L-threonine dehydrogenase by site-directed mutagenesis of cysteine-38.Arch Biochem Biophys. 1998 Oct 15;358(2):211-21. doi: 10.1006/abbi.1998.0845. Arch Biochem Biophys. 1998. PMID: 9784233
Cited by
-
The PduM protein is a structural component of the microcompartments involved in coenzyme B(12)-dependent 1,2-propanediol degradation by Salmonella enterica.J Bacteriol. 2012 Apr;194(8):1912-8. doi: 10.1128/JB.06529-11. Epub 2012 Feb 17. J Bacteriol. 2012. PMID: 22343294 Free PMC article.
-
Co-translational Installation of Posttranslational Modifications by Non-canonical Amino Acid Mutagenesis.Chembiochem. 2023 May 2;24(9):e202300039. doi: 10.1002/cbic.202300039. Epub 2023 Mar 30. Chembiochem. 2023. PMID: 36853967 Free PMC article.
-
Interactions between the termini of lumen enzymes and shell proteins mediate enzyme encapsulation into bacterial microcompartments.Proc Natl Acad Sci U S A. 2012 Sep 11;109(37):14995-5000. doi: 10.1073/pnas.1207516109. Epub 2012 Aug 27. Proc Natl Acad Sci U S A. 2012. PMID: 22927404 Free PMC article.
-
Crystal structures of Staphylococcus epidermidis mevalonate diphosphate decarboxylase bound to inhibitory analogs reveal new insight into substrate binding and catalysis.J Biol Chem. 2011 Jul 8;286(27):23900-10. doi: 10.1074/jbc.M111.242016. Epub 2011 May 11. J Biol Chem. 2011. PMID: 21561869 Free PMC article.
-
The l-Thr Kinase/l-Thr-Phosphate Decarboxylase (CobD) Enzyme from Methanosarcina mazei Gö1 Contains Metallocenters Needed for Optimal Activity.Biochemistry. 2019 Jul 30;58(30):3260-3279. doi: 10.1021/acs.biochem.9b00268. Epub 2019 Jul 17. Biochemistry. 2019. PMID: 31268299 Free PMC article.
References
-
- Banerjee R. (ed) (1999) Chemistry and Biochemistry of B12, John Wiley and Sons, New York
-
- Schneider Z., Stroinski A. (1987) Comprehensive B12: Chemistry, Biochemistry, Nutrition, Ecology, Medicine, De Gruyter, Berlin
-
- Roth J. R., Lawrence J. G., Bobik T. A. (1996) Annu. Rev. Microbiol. 50, 137–181 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

