Crystal structure of the complex between Pseudomonas effector AvrPtoB and the tomato Pto kinase reveals both a shared and a unique interface compared with AvrPto-Pto
- PMID: 19509331
- PMCID: PMC2714939
- DOI: 10.1105/tpc.109.066878
Crystal structure of the complex between Pseudomonas effector AvrPtoB and the tomato Pto kinase reveals both a shared and a unique interface compared with AvrPto-Pto
Abstract
Resistance to bacterial speck disease in tomato (Solanum lycopersicum) is activated upon recognition by the host Pto kinase of either one of two sequence-unrelated effector proteins, AvrPto or AvrPtoB, from Pseudomonas syringae pv tomato (Pst). Pto induces Pst immunity by acting in concert with the Prf protein. The recently reported structure of the AvrPto-Pto complex revealed that interaction of AvrPto with Pto appears to relieve an inhibitory effect of Pto, allowing Pto to activate Prf. Here, we present the crystal structure of the Pto binding domain of AvrPtoB (residues 121 to 205) at a resolution of 1.9A and of the AvrPtoB(121-205)-Pto complex at a resolution of 3.3 A. AvrPtoB(121-205) exhibits a tertiary fold that is completely different from that of AvrPto, and its conformation remains largely unchanged upon binding to Pto. In common with AvrPto-Pto, the AvrPtoB-Pto complex relies on two interfaces. One of these interfaces is similar in both complexes, although the primary amino acid sequences from the two effector proteins are very different. Amino acid substitutions in Pto at the other interface disrupt the interaction of AvrPtoB-Pto but not that of AvrPto-Pto. Interestingly, substitutions in Pto affecting this unique interface also cause Pto to induce Prf-dependent host cell death independently of either effector protein.
Figures

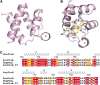




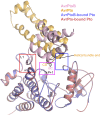
Comment in
-
The tomato Pto kinase uses shared and unique surfaces to recognize divergent avirulence proteins.Plant Cell. 2009 Jun;21(6):1623. doi: 10.1105/tpc.109.210612. Epub 2009 Jun 19. Plant Cell. 2009. PMID: 19542294 Free PMC article. No abstract available.
References
-
- Bernal, A.J., Pan, Q., Pollack, J., Rose, L., Kozik, A., Willits, N., Luo, Y., Guittet, M., Kochetkova, E., and Michelmore, R.W. (2005). Functional analysis of the plant disease resistance gene Pto using DNA shuffling. J. Biol. Chem. 280 23073–23083. - PubMed
-
- Brünger, A.T., et al. (1998). Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 50 905–921. - PubMed
-
- Chang, J.H., Tobias, C.M., Staskawicz, B.J., and Michelmore, R.W. (2001). Functional studies of the bacterial avirulence protein AvrPto by mutational analysis. Mol. Plant Microbe Interact. 14 451–459. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

