Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation
- PMID: 19513119
- PMCID: PMC2688759
- DOI: 10.1371/journal.pone.0005822
Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation
Abstract
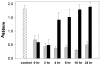
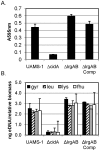
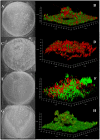
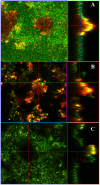
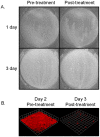
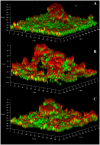
Recent studies have demonstrated a role for Staphylococcus aureus cidA-mediated cell lysis and genomic DNA release in biofilm adherence. The current study extends these findings by examining both temporal and additional genetic factors involved in the control of genomic DNA release and degradation during biofilm maturation. Cell lysis and DNA release were found to be critical for biofilm attachment during the initial stages of development and the released DNA (eDNA) remained an important matrix component during biofilm maturation. This study also revealed that an lrgAB mutant exhibits increased biofilm adherence and matrix-associated eDNA consistent with its proposed role as an inhibitor of cidA-mediated lysis. In flow-cell assays, both cid and lrg mutations had dramatic effects on biofilm maturation and tower formation. Finally, staphylococcal thermonuclease was shown to be involved in biofilm development as a nuc mutant formed a thicker biofilm containing increased levels of matrix-associated eDNA. Together, these findings suggest a model in which the opposing activities of the cid and lrg gene products control cell lysis and genomic DNA release during biofilm development, while staphylococcal thermonuclease functions to degrade the eDNA, possibly as a means to promote biofilm dispersal.
Conflict of interest statement
Figures
References
-
- Costerton JW, Cheng KJ, Geesey GG, Ladd TI, Nickel JC, et al. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41:435–464. - PubMed
-
- Vuong C, Otto M. The biofilm exopolysaccharide polysaccharide intercellular adhesion–a molecular and biochemical approach. Methods Mol Biol. 2008;431:97–106. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

