The expanding world of methylotrophic metabolism
- PMID: 19514844
- PMCID: PMC2827926
- DOI: 10.1146/annurev.micro.091208.073600
The expanding world of methylotrophic metabolism
Abstract
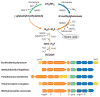
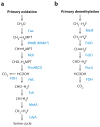
In the past few years, the field of methylotrophy has undergone a significant transformation in terms of discovery of novel types of methylotrophs, novel modes of methylotrophy, and novel metabolic pathways. This time has also been marked by the resolution of long-standing questions regarding methylotrophy and the challenge of long-standing dogmas. This chapter is not intended to provide a comprehensive review of metabolism of methylotrophic bacteria. Instead we focus on significant recent discoveries that are both refining and transforming the current understanding of methylotrophy as a metabolic phenomenon. We also review new directions in methylotroph ecology that improve our understanding of the role of methylotrophy in global biogeochemical processes, along with an outlook for the future challenges in the field.
Figures

References
-
- Alber BE, Spanheimer R, Ebenau-Jehle C, Fuchs G. Study of an alternate glyoxylate cycle for acetate assimilation by Rhodobacter sphaeroides. Mol Microbiol. 2006;61:297–309. - PubMed
-
- Anthony C. The Biochemistry of Methylotrophs. London: Academic; 1982.
-
- Anthony C. The quinoprotein dehydrogenases for methanol and glucose. Arch Biochem Biophys. 2004;428:2–9. - PubMed
-
- Baani M, Liesack W. Two isozymes of particulate methane monooxygenase with different methane oxidation kinetics are found in Methylocystis sp. strain SC2. Proc Natl Acad Sci USA. 2008;105:10203–8. Shows that an organism of the Methylocystis genus is capable of low-affinity methane oxidation. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

