Turnover of organelles by autophagy in yeast
- PMID: 19515549
- PMCID: PMC2725217
- DOI: 10.1016/j.ceb.2009.04.015
Turnover of organelles by autophagy in yeast
Abstract
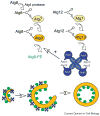
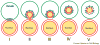
Efficient detection and removal of superfluous or damaged organelles are crucial to maintain cellular homeostasis and to assure cell survival. Growing evidence shows that organelles or parts of them can be removed by selective subtypes of otherwise unselective macroautophagy and microautophagy. This requires both the adaptation of the core autophagic machinery and sophisticated mechanisms to recognize organelles destined for turnover. We review the current knowledge on autophagic removal of peroxisomes, mitochondria, ER and parts of the nucleus with an emphasis on yeasts as a model eukaryote.
Figures
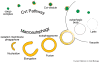

References
-
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. - PubMed
-
- Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. - PubMed
-
- Thumm M, Egner R, Koch B, Schlumpberger M, Straub M, Veenhuis M, Wolf DH. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349:275–280. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

