Viral suppressors of RNA silencing hinder exogenous and endogenous small RNA pathways in Drosophila
- PMID: 19516905
- PMCID: PMC2689938
- DOI: 10.1371/journal.pone.0005866
Viral suppressors of RNA silencing hinder exogenous and endogenous small RNA pathways in Drosophila
Abstract
Background: In plants and insects, RNA interference (RNAi) is the main responder against viruses and shapes the basis of antiviral immunity. Viruses counter this defense by expressing viral suppressors of RNAi (VSRs). While VSRs in Drosophila melanogaster were shown to inhibit RNAi through different modes of action, whether they act on other silencing pathways remained unexplored.
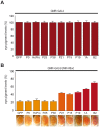
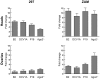
Methodology/principal findings: Here we show that expression of various plant and insect VSRs in transgenic flies does not perturb the Drosophila microRNA (miRNA) pathway; but in contrast, inhibits antiviral RNAi and the RNA silencing response triggered by inverted repeat transcripts, and injection of dsRNA or siRNA. Strikingly, these VSRs also suppressed transposon silencing by endogenous siRNAs (endo-siRNAs).
Conclusions/significance: Our findings identify VSRs as tools to unravel small RNA pathways in insects and suggest a cosuppression of antiviral RNAi and endo-siRNA silencing by viruses during fly infections.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

