Novel genotypes of H9N2 influenza A viruses isolated from poultry in Pakistan containing NS genes similar to highly pathogenic H7N3 and H5N1 viruses
- PMID: 19517011
- PMCID: PMC2690689
- DOI: 10.1371/journal.pone.0005788
Novel genotypes of H9N2 influenza A viruses isolated from poultry in Pakistan containing NS genes similar to highly pathogenic H7N3 and H5N1 viruses
Abstract
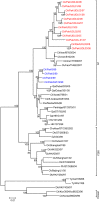
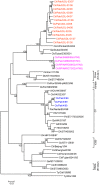
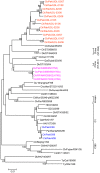
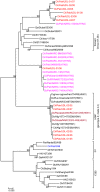
The impact of avian influenza caused by H9N2 viruses in Pakistan is now significantly more severe than in previous years. Since all gene segments contribute towards the virulence of avian influenza virus, it was imperative to investigate the molecular features and genetic relationships of H9N2 viruses prevalent in this region. Analysis of the gene sequences of all eight RNA segments from 12 viruses isolated between 2005 and 2008 was undertaken. The hemagglutinin (HA) sequences of all isolates were closely related to H9N2 viruses isolated from Iran between 2004 and 2007 and contained leucine instead of glutamine at position 226 in the receptor binding pocket, a recognised marker for the recognition of sialic acids linked alpha2-6 to galactose. The neuraminidase (NA) of two isolates contained a unique five residue deletion in the stalk (from residues 80 to 84), a possible indication of greater adaptation of these viruses to the chicken host. The HA, NA, nucleoprotein (NP), and matrix (M) genes showed close identity with H9N2 viruses isolated during 1999 in Pakistan and clustered in the A/Quail/Hong Kong/G1/97 virus lineage. In contrast, the polymerase genes clustered with H9N2 viruses from India, Iran and Dubai. The NS gene segment showed greater genetic diversity and shared a high level of similarity with NS genes from either H5 or H7 subtypes rather than with established H9N2 Eurasian lineages. These results indicate that during recent years the H9N2 viruses have undergone extensive genetic reassortment which has led to the generation of H9N2 viruses of novel genotypes in the Indian sub-continent. The novel genotypes of H9N2 viruses may play a role in the increased problems observed by H9N2 to poultry and reinforce the continued need to monitor H9N2 infections for their zoonotic potential.
Conflict of interest statement
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

