Can systems biology understand pathway activation? Gene expression signatures as surrogate markers for understanding the complexity of pathway activation
- PMID: 19517027
- PMCID: PMC2694555
- DOI: 10.2174/138920208785133235
Can systems biology understand pathway activation? Gene expression signatures as surrogate markers for understanding the complexity of pathway activation
Abstract
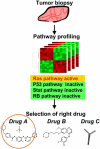
Cancer is thought to be caused by a sequence of multiple genetic and epigenetic alterations which occur in one or more of the genes controlling cell cycle progression and signaling transduction. The complexity of carcinogenic mechanisms leads to heterogeneity in molecular phenotype, pathology, and prognosis of cancers.Genome-wide mutational analysis of cancer genes in individual tumors is the most direct way to elucidate the complex process of disease progression, although such high-throughput sequencing technologies are not yet fully developed. As a surrogate marker for pathway activation analysis, expression profiling using microarrays has been successfully applied for the classification of tumor types, stages of tumor progression, or in some cases, prediction of clinical outcomes. However, the biological implication of those gene expression signatures is often unclear. Systems biological approaches leverage the signature genes as a representation of changes in signaling pathways, instead of interpreting the relevance between each gene and phenotype. This approach, which can be achieved by comparing the gene set or the expression profile with those of reference experiments in which a defined pathway is modulated, will improve our understanding of cancer classification, clinical outcome, and carcinogenesis. In this review, we will discuss recent studies on the development of expression signatures to monitor signaling pathway activities and how these signatures can be used to improve the identification of responders to anticancer drugs.
Keywords: Expression signature; cancer therapy; drug discovery; signaling pathway; systems biology..
Figures
References
-
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. - PubMed
-
- Parsons DW, Wang TL, Samuels Y, Bardelli A, Cummins JM, DeLong L, Silliman N, Ptak J, Szabo S, Willson JKV, Markowitz S, Kinzler K, Vogelstein B, Lengauer C, Velculescu VE. Colorectal cancer - Mutations in a signalling pathway. Nature. 2005;436:792. - PubMed
-
- Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, Silliman N, Szabo S, Dezso Z, Ustyanksky V, Nikolskaya T, Nikolsky Y, Karchin R, Wilson PA, Kaminker JS, Zhang ZM, Croshaw R, Willis J, Dawson D, Shipitsin M, Willson JKV, Sukumar S, Polyak K, Park BH, Pethiyagoda CL, Pant PVK, Ballinger DG, Sparks AB, Hartigan J, Smith DR, Suh E, Papadopoulos N, Buckhaults P, Markowitz SD, Parmigiani G, Kinzler KW, Velculescu VE, Vogelstein B. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. - PubMed
-
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Edkins S, O'Meara S, Vastrik I, Schmidt EE, Avis T, Barthorpe S, Bhamra G, Buck G, Choudhury B, Clements J, Cole J, Dicks E, Forbes S, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jenkinson A, Jones D, Menzies A, Mironenko T, Perry J, Raine K, Richardson D, Shepherd R, Small A, Tofts C, Varian J, Webb T, West S, Widaa S, Yates A, Cahill DP, Louis DN, Goldstraw P, Nicholson AG, Brasseur F, Looijenga L, Weber BL, Chiew YE, Defazio A, Greaves MF, Green AR, Campbell P, Birney E, Easton DF, Chenevix-Trench G, Tan MH, Khoo SK, Teh BT, Yuen ST, Leung SY, Wooster R, Futreal PA, Stratton MR. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. - PMC - PubMed
-
- Thomas RK, Baker AC, DeBiasi RM, Winckler W, LaFramboise T, Lin WM, Wang M, Feng W, Zander T, MacConnaill LE, Lee JC, Nicoletti R, Hatton C, Goyette M, Girard L, Majmudar K, Ziaugra L, Wong KK, Gabriel S, Beroukhim R, Peyton M, Barretina J, Dutt A, Emery C, Greulich H, Shah K, Sasaki H, Gazdar A, Minna J, Armstrong SA, Mellinghoff IK, Hodi FS, Dranoff G, Mischel PS, Cloughesy TF, Nelson SF, Liau LM, Mertz K, Rubin MA, Moch H, Loda M, Catalona W, Fletcher J, Signoretti S, Kaye F, Anderson KC, Demetri GD, Dummer R, Wagner S, Herlyn M, Sellers WR, Meyerson M, Garraway LA. High-throughput oncogene mutation profiling in human cancer. Nat. Genet. 2007;39:347–351. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
