Structural and functional divergence within the Dim1/KsgA family of rRNA methyltransferases
- PMID: 19520088
- PMCID: PMC2753216
- DOI: 10.1016/j.jmb.2009.06.015
Structural and functional divergence within the Dim1/KsgA family of rRNA methyltransferases
Abstract
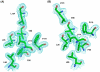
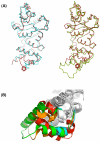
The enzymes of the KsgA/Dim1 family are universally distributed throughout all phylogeny; however, structural and functional differences are known to exist. The well-characterized function of these enzymes is to dimethylate two adjacent adenosines of the small ribosomal subunit in the normal course of ribosome maturation, and the structures of KsgA from Escherichia coli and Dim1 from Homo sapiens and Plasmodium falciparum have been determined. To this point, no examples of archaeal structures have been reported. Here, we report the structure of Dim1 from the thermophilic archaeon Methanocaldococcus jannaschii. While it shares obvious similarities with the bacterial and eukaryotic orthologs, notable structural differences exist among the three members, particularly in the C-terminal domain. Previous work showed that eukaryotic and archaeal Dim1 were able to robustly complement for KsgA in E. coli. Here, we repeated similar experiments to test for complementarity of archaeal Dim1 and bacterial KsgA in Saccharomyces cerevisiae. However, neither the bacterial nor the archaeal ortholog could complement for the eukaryotic Dim1. This might be related to the secondary, non-methyltransferase function that Dim1 is known to play in eukaryotic ribosomal maturation. To further delineate regions of the eukaryotic Dim1 critical to its function, we created and tested KsgA/Dim1 chimeras. Of the chimeras, only one constructed with the N-terminal domain from eukaryotic Dim1 and the C-terminal domain from archaeal Dim1 was able to complement, suggesting that eukaryotic-specific Dim1 function resides in the N-terminal domain also, where few structural differences are observed between members of the KsgA/Dim1 family. Future work is required to identify those determinants directly responsible for Dim1 function in ribosome biogenesis. Finally, we have conclusively established that none of the methyl groups are critically important to growth in yeast under standard conditions at a variety of temperatures.
Figures




References
-
- Poldermans B, Roza L, Van Knippenberg PH. Studies on the function of two adjacent N6,N6-dimethyladenosines near the 3′ end of 16 S ribosomal RNA of Escherichia coli. III. Purification and properties of the methylating enzyme and methylase-30 S interactions. J. Biol. Chem. 1979;254:9094–9100. - PubMed
-
- van Buul CP, van Knippenberg PH. Nucleotide sequence of the ksgA gene of Escherichia coli: comparison of methyltransferases effecting dimethylation of adenosine in ribosomal RNA. Gene. 1985;38:65–72. - PubMed
-
- Van Buul CP, Damm JB, Van Knippenberg PH. Kasugamycin resistant mutants of Bacillus stearothermophilus lacking the enzyme for the methylation of two adjacent adenosines in 16S ribosomal RNA. Mol. Gen. Genet. 1983;189:475–478. - PubMed
-
- Lontaine D, Delcour J, Glasser AL, Desgres J, Vandenhaute J. The DIM1 gene responsible for the conserved m6(2)Am6(2)A dimethylation in the 3′-terminal loop of 18 S rRNA is essential in yeast. J. Mol. Biol. 1994;241:492–497. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

