Src homology 2 domain-containing inositol-5-phosphatase and CCAAT enhancer-binding protein beta are targeted by miR-155 in B cells of Emicro-MiR-155 transgenic mice
- PMID: 19520806
- PMCID: PMC2727407
- DOI: 10.1182/blood-2009-05-220814
Src homology 2 domain-containing inositol-5-phosphatase and CCAAT enhancer-binding protein beta are targeted by miR-155 in B cells of Emicro-MiR-155 transgenic mice
Abstract
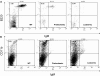
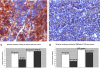
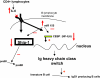
We showed that Emicro-MiR-155 transgenic mice develop acute lymphoblastic leukemia/high-grade lymphoma. Most of these leukemias start at approximately 9 months irrespective of the mouse strain. They are preceded by a polyclonal pre-B-cell proliferation, have variable clinical presentation, are transplantable, and develop oligo/monoclonal expansion. In this study, we show that in these transgenic mice the B-cell precursors have the highest MiR-155 transgene expression and are at the origin of the leukemias. We determine that Src homology 2 domain-containing inositol-5-phosphatase (SHIP) and CCAAT enhancer-binding protein beta (C/EBPbeta), 2 important regulators of the interleukin-6 signaling pathway, are direct targets of MiR-155 and become gradually more down-regulated in the leukemic than in the preleukemic mice. We hypothesize that miR-155, by down-modulating Ship and C/EBPbeta, initiates a chain of events that leads to the accumulation of large pre-B cells and acute lymphoblastic leukemia/high-grade lymphoma.
Figures

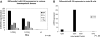


References
-
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. - PubMed
-
- Calin GA, Ferracin A, Cimmino A, et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2006;353:1793–1801. - PubMed
-
- Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

