The Thermoplasma acidophilum LplA-LplB complex defines a new class of bipartite lipoate-protein ligases
- PMID: 19520844
- PMCID: PMC2755856
- DOI: 10.1074/jbc.M109.015016
The Thermoplasma acidophilum LplA-LplB complex defines a new class of bipartite lipoate-protein ligases
Abstract
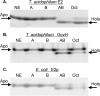
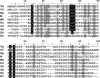
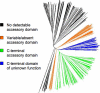
Lipoic acid is a covalently bound cofactor found throughout the domains of life that is required for aerobic metabolism of 2-oxoacids and for C(1) metabolism. Utilization of exogenous lipoate is catalyzed by a ligation reaction that proceeds via a lipoyl-adenylate intermediate to attach the cofactor to the epsilon-amino group of a conserved lysine residue of protein lipoyl domains. The lipoyl ligases of demonstrated function have a large N-terminal catalytic domain and a small C-terminal accessory domain. Half of the members of the LplA family detected in silico have only the large catalytic domain. Two x-ray structures of the Thermoplasma acidophilum LplA structure have been reported, although the protein was reported to lack ligase activity. McManus et al. (McManus, E., Luisi, B. F., and Perham, R. N. (2006) J. Mol. Biol. 356, 625-637) hypothesized that the product of an adjacent gene was also required for ligase activity. We have shown this to be the case and have named the second protein, LplB. We found that complementation of Escherichia coli strains lacking lipoate ligase with T. acidophilum LplA was possible only when LplB was also present. LplA had no detectable ligase activity in vitro in the absence of LplB. Moreover LplA and LplB were shown to interact and were purified as a heterodimer. LplB was required for lipoyl-adenylate formation but was not required for transfer of the lipoyl moiety of lipoyl-adenylate to acceptor proteins. Surveys of sequenced genomes show that most lipoyl ligases of the kingdom Archaea are heterodimeric. We propose that the presence of an accessory domain provides a diagnostic to distinguish lipoyl ligase homologues from other members of the lipoate/biotin attachment enzyme family.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

