A genome-wide screen for regulators of TORC1 in response to amino acid starvation reveals a conserved Npr2/3 complex
- PMID: 19521502
- PMCID: PMC2686269
- DOI: 10.1371/journal.pgen.1000515
A genome-wide screen for regulators of TORC1 in response to amino acid starvation reveals a conserved Npr2/3 complex
Abstract
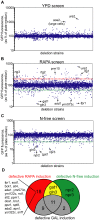
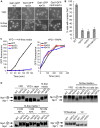
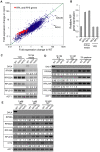
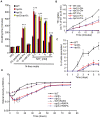
TORC1 is a central regulator of cell growth in response to amino acid availability, yet little is known about how it is regulated. Here, we performed a reverse genetic screen in yeast for genes necessary to inactivate TORC1. The screen consisted of monitoring the expression of a TORC1 sensitive GFP-based transcriptional reporter in all yeast deletion strains using flow cytometry. We find that in response to amino acid starvation, but not to carbon starvation or rapamycin treatment, cells lacking NPR2 and NPR3 fail to fully (1) activate transcription factors Gln3/Gat1, (2) dephosphorylate TORC1 effector Npr1, and (3) repress ribosomal protein gene expression. Both mutants show proliferation defects only in media containing a low quality nitrogen source, such as proline or ammonia, whereas no defects are evident when cells are grown in the presence of glutamine or peptone mixture. Proliferation defects in npr2Delta and npr3Delta cells can be completely rescued by artificially inhibiting TORC1 by rapamycin, demonstrating that overactive TORC1 in both strains prevents their ability to adapt to an environment containing a low quality nitrogen source. A biochemical purification of each demonstrates that Npr2 and Npr3 form a heterodimer, and this interaction is evolutionarily conserved since the human homologs of NPR2 and NPR3 (NPRL2 and NPRL3, respectively) also co-immunoprecipitate. We conclude that, in yeast, the Npr2/3 complex mediates an amino acid starvation signal to TORC1.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. - PubMed
-
- Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. - PubMed
-
- Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Current biology. 2004;14:1296–1302. - PubMed
-
- Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Molecular Cell. 2002;10:457–468. - PubMed
-
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

