MicroRNA expression profiling in mild asthmatic human airways and effect of corticosteroid therapy
- PMID: 19521514
- PMCID: PMC2690402
- DOI: 10.1371/journal.pone.0005889
MicroRNA expression profiling in mild asthmatic human airways and effect of corticosteroid therapy
Abstract
Background: Asthma is a common disease characterised by reversible airflow obstruction, bronchial hyperresponsiveness and chronic inflammation, which is commonly treated using corticosteroids such as budesonide. MicroRNAs (miRNAs) are a recently identified family of non-protein encoding genes that regulate protein translation by a mechanism entitled RNA interference. Previous studies have shown lung-specific miRNA expression profiles, although their importance in regulating gene expression is unresolved. We determined whether miRNA expression was differentially expressed in mild asthma and the effect of corticosteroid treatment.
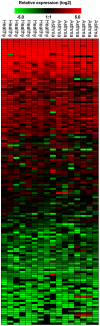
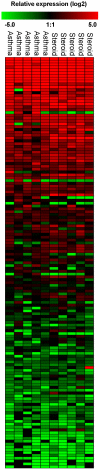
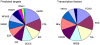
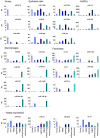
Methodology/principal findings: We have examined changes in miRNA using a highly sensitive RT-PCR based approach to measure the expression of 227 miRNAs in airway biopsies obtained from normal and mild asthmatic patients. We have also determined whether the anti-inflammatory action of corticosteroids are mediated through miRNAs by determining the profile of miRNA expression in mild asthmatics, before and following 1 month twice daily treatment with inhaled budesonide. Furthermore, we have analysed the expression of miRNAs from individual cell populations from the airway and lung. We found no significant difference in the expression of 227 miRNAs in the airway biopsies obtained from normal and mild asthmatic patients. In addition, despite improved lung function, we found no significant difference in the miRNA expression following one month treatment with the corticosteroid, budesonide. However, analysis of bronchial and alveolar epithelial cells, airway smooth muscle cells, alveolar macrophages and lung fibroblasts demonstrate a miRNA expression profile that is specific to individual cell types and demonstrates the complex cellular heterogeneity within whole tissue samples.
Conclusions: Changes in miRNA expression do not appear to be involved in the development of a mild asthmatic phenotype or in the anti-inflammatory action of the corticosteroid budesonide.
Conflict of interest statement
Figures
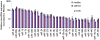
References
-
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. - PubMed
-
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. - PubMed
-
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, et al. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. - PubMed
-
- Tang G. siRNA and miRNA: an insight into RISCs. Trends Biochem Sci. 2005;30:106–114. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

