Biochemical and structural characterization of the human TL1A ectodomain
- PMID: 19522538
- PMCID: PMC2790920
- DOI: 10.1021/bi900031w
Biochemical and structural characterization of the human TL1A ectodomain
Abstract
TNF-like 1A (TL1A) is a newly described member of the TNF superfamily that is directly implicated in the pathogenesis of autoimmune diseases, including inflammatory bowel disease, atherosclerosis, and rheumatoid arthritis. We report the crystal structure of the human TL1A extracellular domain at a resolution of 2.5 A, which reveals a jelly-roll fold typical of the TNF superfamily. This structural information, in combination with complementary mutagenesis and biochemical characterization, provides insights into the binding interface and the specificity of the interactions between TL1A and the DcR3 and DR3 receptors. These studies suggest that the mode of interaction between TL1A and DcR3 differs from other characterized TNF ligand/receptor complexes. In addition, we have generated functional TL1A mutants with altered disulfide bonding capability that exhibit enhanced solution properties, which will facilitate the production of materials for future cell-based and whole animal studies. In summary, these studies provide insights into the structure and function of TL1A and provide the basis for the rational manipulation of its interactions with cognate receptors.
Figures

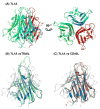
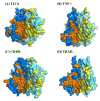




References
-
- Migone TS, Zhang J, Luo X, Zhuang L, Chen C, Hu B, Hong JS, Perry JW, Chen SF, Zhou JX, Cho YH, Ullrich S, Kanakaraj P, Carrell J, Boyd E, Olsen HS, Hu G, Pukac L, Liu D, Ni J, Kim S, Gentz R, Feng P, Moore PA, Ruben SM, Wei P. TL1A is a TNF-like ligand for DR3 and TR6/DcR3 and functions as a T cell costimulator. Immunity. 2002;16:479–492. - PubMed
-
- Chew LJ, Pan H, Yu J, Tian S, Huang WQ, Zhang JY, Pang S, Li LY. A novel secreted splice variant of vascular endothelial cell growth inhibitor. Faseb J. 2002;16:742–744. - PubMed
-
- Yang CR, Hsieh SL, Teng CM, Ho FM, Su WL, Lin WW. Soluble decoy receptor 3 induces angiogenesis by neutralization of TL1A, a cytokine belonging to tumor necrosis factor superfamily and exhibiting angiostatic action. Cancer Res. 2004;64:1122–1129. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

