The ubiquitin-proteasome system in cancer, a major player in DNA repair. Part 1: post-translational regulation
- PMID: 19522845
- PMCID: PMC4516461
- DOI: 10.1111/j.1582-4934.2009.00824.x
The ubiquitin-proteasome system in cancer, a major player in DNA repair. Part 1: post-translational regulation
Abstract
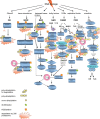
DNA repair is a fundamental cellular function, indispensable for cell survival, especially in conditions of exposure to environmental or pharmacological effectors of DNA damage. The regulation of this function requires a flexible machinery to orchestrate the reversal of harmful DNA lesions by making use of existing proteins as well as inducible gene products. The accumulation of evidence for the involvement of ubiquitin-proteasome system (UPS) in DNA repair pathways, that is reviewed here, has expanded its role from a cellular waste disposal basket to a multi-dimensional regulatory system. This review is the first of two that attempt to illustrate the nature and interactions of all different DNA repair pathways where UPS is demonstrated to be involved, with special focus on cancer- and chemotherapy-related DNA-damage repair. In this first review, we will be presenting the proteolytic and non-proteolytic roles of UPS in the post-translational regulation of DNA repair proteins, while the second review will focus on the UPS-dependent transcriptional response of DNA repair after DNA damage and stress.
Figures
References
-
- Hansen WK, Kelley MR. Review of mammalian DNA repair and translational implications. J Pharmacol Exp Ther. 2000;295:1–9. - PubMed
-
- Christmann M, Tomicic MT, Roos WP, et al. Mechanisms of human DNA repair: an update. Toxicology. 2003;193:3–34. - PubMed
-
- Wood RD, Mitchell M, Lindahl T. Human DNA repair genes. Mutat Res. 2005;577:275–83. - PubMed
-
- Pourquier P. La réparation de l’ ADN, cible potentielle d’un développement thérapeutique en cancérologie. Bull Cancer. 2006:124–44. ; hors série:
-
- Sánchez-Pérez I. DNA inhibitors in cancer treatment. Clin Transl Oncol. 2006;8:642–6. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

