Maintenance of genetic variation in plants and pathogens involves complex networks of gene-for-gene interactions
- PMID: 19523099
- PMCID: PMC6640458
- DOI: 10.1111/j.1364-3703.2009.00544.x
Maintenance of genetic variation in plants and pathogens involves complex networks of gene-for-gene interactions
Abstract
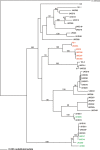
The RPP13 [recognition of Hyaloperonospora arabidopsidis (previously known as Peronospora parasitica)] resistance (R) gene in Arabidopsis thaliana exhibits the highest reported level of sequence diversity among known R genes. Consistent with a co-evolutionary model, the matching effector protein ATR13 (A. thaliana-recognized) from H. arabidopsidis reveals extreme levels of allelic diversity. We isolated 23 new RPP13 sequences from a UK metapopulation, giving a total of 47 when combined with previous studies. We used these in functional studies of the A. thaliana accessions for their resistance response to 16 isolates of H. arabidopsidis. We characterized the molecular basis of recognition by the expression of the corresponding ATR13 genes from these 16 isolates in these host accessions. This allowed the determination of which alleles of RPP13 were responsible for pathogen recognition and whether recognition was dependent on the RPP13/ATR13 combination. Linking our functional studies with phylogenetic analysis, we determined that: (i) the recognition of ATR13 is mediated by alleles in just a single RPP13 clade; (ii) RPP13 alleles in other clades have evolved the ability to detect other pathogen ATR protein(s); and (iii) at least one gene, unlinked to RPP13 in A. thaliana, detects a different subgroup of ATR13 alleles.
Figures

References
-
- Allen, R.L. , Bittner‐Eddy, P.D. , Grenville‐Briggs, L.J. , Meitz, J.C. , Rehmany, A.P. , Rose, L.E. and Beynon, J.L. (2004) Host–parasite coevolutionary conflict between Arabidopsis and downy mildew. Science, 306, 1957–1960. - PubMed
-
- Axtell, M.J. and Staskawicz, B.J. (2003) Initiation of RPS2‐specified disease resistance in Arabidopsis is coupled to the AvrRpt2‐directed elimination of RIN4. Cell, 112, 369–377. - PubMed
-
- Van Der Biezen, E.A. and Jones, J.D.G. (1998) Plant disease resistance proteins and the gene‐for‐gene concept. Trends Biochem. Sci. 23, 454–456. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

