BASL controls asymmetric cell division in Arabidopsis
- PMID: 19523675
- PMCID: PMC4105981
- DOI: 10.1016/j.cell.2009.04.018
BASL controls asymmetric cell division in Arabidopsis
Abstract
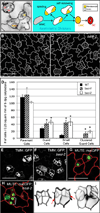
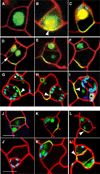
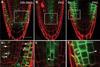
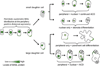
Development in multicellular organisms requires the organized generation of differences. A universal mechanism for creating such differences is asymmetric cell division. In plants, as in animals, asymmetric divisions are correlated with the production of cellular diversity and pattern; however, structural constraints imposed by plant cell walls and the absence of homologs of known animal or fungal cell polarity regulators necessitates that plants utilize new molecules and mechanisms to create asymmetries. Here, we identify BASL, a novel regulator of asymmetric divisions in Arabidopsis. In asymmetrically dividing stomatal-lineage cells, BASL accumulates in a polarized crescent at the cell periphery before division, and then localizes differentially to the nucleus and a peripheral crescent in self-renewing cells and their sisters after division. BASL presence at the cell periphery is critical for its function, and we propose that BASL represents a plant-specific solution to the challenge of asymmetric cell division.
Figures



Similar articles
-
Evolution of polarity protein BASL and the capacity for stomatal lineage asymmetric divisions.Curr Biol. 2022 Jan 24;32(2):329-337.e5. doi: 10.1016/j.cub.2021.11.013. Epub 2021 Nov 29. Curr Biol. 2022. PMID: 34847354
-
The Arabidopsis stomatal polarity protein BASL mediates distinct processes before and after cell division to coordinate cell size and fate asymmetries.Development. 2021 Sep 15;148(18):dev199919. doi: 10.1242/dev.199919. Epub 2021 Sep 27. Development. 2021. PMID: 34463761 Free PMC article.
-
Fine-scale dissection of the subdomains of polarity protein BASL in stomatal asymmetric cell division.J Exp Bot. 2016 Sep;67(17):5093-103. doi: 10.1093/jxb/erw274. Epub 2016 Jul 15. J Exp Bot. 2016. PMID: 27422992 Free PMC article.
-
Division polarity in developing stomata.Curr Opin Plant Biol. 2012 Dec;15(6):585-92. doi: 10.1016/j.pbi.2012.09.013. Epub 2012 Oct 5. Curr Opin Plant Biol. 2012. PMID: 23044038 Review.
-
Stomatal patterning and development.Curr Top Dev Biol. 2010;91:267-97. doi: 10.1016/S0070-2153(10)91009-0. Curr Top Dev Biol. 2010. PMID: 20705185 Review.
Cited by
-
Opposite polarity programs regulate asymmetric subsidiary cell divisions in grasses.Elife. 2022 Dec 20;11:e79913. doi: 10.7554/eLife.79913. Elife. 2022. PMID: 36537077 Free PMC article.
-
The importance of being edgy: cell geometric edges as an emerging polar domain in plant cells.J Microsc. 2020 Jun;278(3):123-131. doi: 10.1111/jmi.12847. Epub 2019 Nov 28. J Microsc. 2020. PMID: 31755561 Free PMC article. Review.
-
SPEECHLESS integrates brassinosteroid and stomata signalling pathways.Nat Cell Biol. 2012 Apr 1;14(5):548-54. doi: 10.1038/ncb2471. Nat Cell Biol. 2012. PMID: 22466366
-
Polarly localized WPR proteins interact with PAN receptors and the actin cytoskeleton during maize stomatal development.Plant Cell. 2023 Jan 2;35(1):469-487. doi: 10.1093/plcell/koac301. Plant Cell. 2023. PMID: 36227066 Free PMC article.
-
Stomatal development in the context of epidermal tissues.Ann Bot. 2021 Jul 30;128(2):137-148. doi: 10.1093/aob/mcab052. Ann Bot. 2021. PMID: 33877316 Free PMC article. Review.
References
-
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. - PubMed
-
- Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, Friml J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. - PubMed
-
- Bergmann DC, Sack FD. Stomatal development. Annu. Rev. Plant Biol. 2007;58:163–181. - PubMed
-
- Bergmann DC, Lukowitz W, Somerville CR. Stomatal development and pattern controlled by a MAPKK kinase. Science. 2004;304:1494–1497. - PubMed
-
- Betschinger J, Mechtler K, Knoblich JA. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in drosophila neural stem cells. Cell. 2006;124:1241–1253. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

