The P22 tail machine at subnanometer resolution reveals the architecture of an infection conduit
- PMID: 19523897
- PMCID: PMC2714705
- DOI: 10.1016/j.str.2009.04.006
The P22 tail machine at subnanometer resolution reveals the architecture of an infection conduit
Abstract
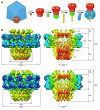
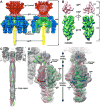
The portal channel is a key component in the life cycle of bacteriophages and herpesviruses. The bacteriophage P22 portal is a 1 megadalton dodecameric oligomer of gp1 that plays key roles in capsid assembly, DNA packaging, assembly of the infection machinery, and DNA ejection. The portal is the nucleation site for the assembly of 39 additional subunits generated from multiple copies of four gene products (gp4, gp10, gp9, and gp26), which together form the multifunctional tail machine. These components are organized with a combination of 12-fold (gp1, gp4), 6-fold (gp10, trimers of gp9), and 3-fold (gp26, gp9) symmetry. Here we present the 3-dimensional structures of the P22 assembly-naive portal formed from expressed subunits (gp1) and the intact tail machine purified from infectious virions. The assembly-naive portal structure exhibits a striking structural similarity to the structures of the portal proteins of SPP1 and phi29 derived from X-ray crystallography.
Figures
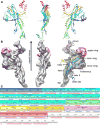
References
-
- Agirrezabala X, Martin-Benito J, Valle M, Gonzalez JM, Valencia A, Valpuesta JM, Carrascosa JL. Structure of the connector of bacteriophage T7 at 8A resolution: structural homologies of a basic component of a DNA translocating machinery. Journal of molecular biology. 2005b;347(5):895–902. - PubMed
-
- Bazinet C, Benbasat J, King J, Carazo JM, Carrascosa JL. Purification and organization of the gene 1 portal protein required for phage P22 DNA packaging. Biochemistry. 1988;27(6):1849–1856. - PubMed
-
- Bhardwaj A, Olia AS, Walker-Kopp N, Cingolani G. Domain organization and polarity of tail needle GP26 in the portal vertex structure of bacteriophage P22. Journal of molecular biology. 2007;371(2):374–387. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

