PSI-2: structural genomics to cover protein domain family space
- PMID: 19523904
- PMCID: PMC2920419
- DOI: 10.1016/j.str.2009.03.015
PSI-2: structural genomics to cover protein domain family space
Abstract
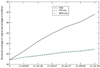
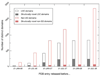
One major objective of structural genomics efforts, including the NIH-funded Protein Structure Initiative (PSI), has been to increase the structural coverage of protein sequence space. Here, we present the target selection strategy used during the second phase of PSI (PSI-2). This strategy, jointly devised by the bioinformatics groups associated with the PSI-2 large-scale production centers, targets representatives from large, structurally uncharacterized protein domain families, and from structurally uncharacterized subfamilies in very large and diverse families with incomplete structural coverage. These very large families are extremely diverse both structurally and functionally, and are highly overrepresented in known proteomes. On the basis of several metrics, we then discuss to what extent PSI-2, during its first 3 years, has increased the structural coverage of genomes, and contributed structural and functional novelty. Together, the results presented here suggest that PSI-2 is successfully meeting its objectives and provides useful insights into structural and functional space.
Figures





References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. - PubMed
-
- Aravind L, Anantharaman V, Koonin EV. Monophyly of class I aminoacyl tRNA synthetase, USPA, ETFP, photolyase, and PP-ATPase nucleotide-binding domains: implications for protein evolution in the RNA. Proteins. 2002;48:1–14. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

