Retinoblastoma has properties of a cone precursor tumor and depends upon cone-specific MDM2 signaling
- PMID: 19524506
- PMCID: PMC5659855
- DOI: 10.1016/j.cell.2009.03.051
Retinoblastoma has properties of a cone precursor tumor and depends upon cone-specific MDM2 signaling
Abstract
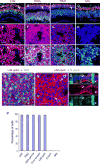
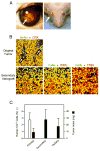
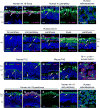
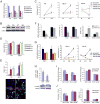
Retinoblastomas result from the inactivation of the RB1 gene and the loss of Rb protein, yet the cell type in which Rb suppresses retinoblastoma and the circuitry that underlies the need for Rb are undefined. Here, we show that retinoblastoma cells express markers of postmitotic cone precursors but not markers of other retinal cell types. We also demonstrate that human cone precursors prominently express MDM2 and N-Myc, that retinoblastoma cells require both of these proteins for proliferation and survival, and that MDM2 is needed to suppress ARF-induced apoptosis in cultured retinoblastoma cells. Interestingly, retinoblastoma cell MDM2 expression was regulated by the cone-specific RXRgamma transcription factor and a human-specific RXRgamma consensus binding site, and proliferation required RXRgamma, as well as the cone-specific thyroid hormone receptor-beta2. These findings provide support for a cone precursor origin of retinoblastoma and suggest that human cone-specific signaling circuitry sensitizes to the oncogenic effects of RB1 mutations.
Figures



Comment in
-
Retinoblastoma, an inside job.Cell. 2009 Jun 12;137(6):992-4. doi: 10.1016/j.cell.2009.05.034. Cell. 2009. PMID: 19524501
References
-
- Abramson DH, Gombos DS. The topography of bilateral retinoblastoma lesions. Retina. 1996;16:232–239. - PubMed
-
- Bibb LC, Holt JK, Tarttelin EE, Hodges MD, Gregory-Evans K, Rutherford A, Lucas RJ, Sowden JC, Gregory-Evans CY. Temporal and spatial expression patterns of the CRX transcription factor and its downstream targets. Critical differences during human and mouse eye development. Hum Mol Genet. 2001;10:1571–1579. - PubMed
-
- Bogenmann E, Lochrie MA, Simon MI. Cone cell-specific genes expressed in retinoblastoma. Science. 1988;240:76–78. - PubMed
-
- Chen D, Livne-bar I, Vanderluit JL, Slack RS, Agochiya M, Bremner R. Cell-specific effects of RB or RB/p107 loss on retinal development implicate an intrinsically death-resistant cell-of-origin in retinoblastoma. Cancer Cell. 2004;5:539–551. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

