LasI/R and RhlI/R quorum sensing in a strain of Pseudomonas aeruginosa beneficial to plants
- PMID: 19525275
- PMCID: PMC2725484
- DOI: 10.1128/AEM.02914-08
LasI/R and RhlI/R quorum sensing in a strain of Pseudomonas aeruginosa beneficial to plants
Abstract
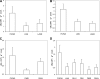
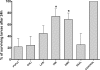
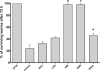
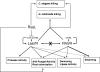
Pseudomonas aeruginosa possesses three quorum-sensing (QS) systems which are key in the expression of a large number of genes, including many virulence factors. Most studies of QS in P. aeruginosa have been performed in clinical isolates and have therefore focused on its role in pathogenicity. P. aeruginosa, however, is regarded as a ubiquitous organism capable of colonizing many different environments and also of establishing beneficial associations with plants. In this study we examined the role of the two N-acyl homoserine lactone systems known as RhlI/R and LasI/R in the environmental rice rhizosphere isolate P. aeruginosa PUPa3. Both the Rhl and Las systems are involved in the regulation of plant growth-promoting traits. The environmental P. aeruginosa PUPa3 is pathogenic in two nonmammalian infection models, and only the double las rhl mutants are attenuated for virulence. In fact it was established that the two QS systems are not hierarchically organized and that they are both important for the colonization of the rice rhizosphere. This is an in-depth genetic and molecular study of QS in an environmental P. aeruginosa strain and highlights several differences with QS regulation in the clinical isolate PAO1.
Figures

References
-
- Alexeyev, M. F. 1999. The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of gram-negative bacteria. BioTechniques 26:824-826, 828. - PubMed
-
- Arevalo-Ferro, C., M. Hentzer, G. Reil, A. Gorg, S. Kjelleberg, M. Givskov, K. Riedel, and L. Eberl. 2003. Identification of quorum-sensing regulated proteins in the opportunistic pathogen Pseudomonas aeruginosa by proteomics. Environ. Microbiol. 5:1350-1369. - PubMed
-
- Berg, G., L. Eberl, and A. Hartmann. 2005. The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environ. Microbiol. 7:1673-1685. - PubMed
-
- Better, M., B. Lewis, D. Corbin, G. Ditta, and D. R. Helinski. 1983. Structural relationships among Rhizobium meliloti symbiotic promoters. Cell 35:479-485. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

