Chromosome congression in the absence of kinetochore fibres
- PMID: 19525938
- PMCID: PMC2895821
- DOI: 10.1038/ncb1890
Chromosome congression in the absence of kinetochore fibres
Abstract
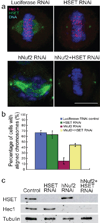
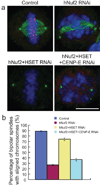
Proper chromosome congression (the process of aligning chromosomes on the spindle) contributes to accurate and faithful chromosome segregation. It is widely accepted that congression requires kinetochore fibres (K-fibres), microtubule bundles that extend from the kinetochores to spindle poles. Here, we demonstrate that chromosomes in human cells co-depleted of HSET (human kinesin-14) and hNuf2 (human Ndc80/Hec1-complex component) can congress to the metaphase plate in the absence of K-fibres. However, the chromosomes are not stably maintained at the metaphase plate under these conditions. Chromosome congression in HSET + hNuf2 co-depleted cells required the plus-end directed motor CENP-E (centromere protein E; kinesin-7 family member), which has been implicated in the gliding of mono-oriented kinetochores alongside adjacent K-fibres. Thus, proper end-on attachment of kinetochores to microtubules is not necessary for chromosome congression. Instead, our data support the idea that congression allows unattached chromosomes to move to the middle of the spindle where they have a higher probability of establishing connections with both spindle poles. These bi-oriented connections are also used to maintain stable chromosome alignment at the spindle equator.
Figures



Comment in
-
Chromosome congression: on the bi-orient express.Nat Cell Biol. 2009 Jul;11(7):787-9. doi: 10.1038/ncb1902. Nat Cell Biol. 2009. PMID: 19532116
References
-
- Maiato H, DeLuca J, Salmon ED, Earnshaw WC. The dynamic kinetochore-microtubule interface. J. Cell Sci. 2004;117:5461–5477. - PubMed
-
- Walczak CE, Heald R. Mechanisms of mitotic spindle assembly and function. Int. Re.v Cytol. 2008;265:111–158. - PubMed
-
- Manning AL, Compton DA. Mechanisms of spindle-pole organization are influenced by kinetochore activity in mammalian cells. Curr. Biol. 2007;17:260–265. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

