Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci
- PMID: 19525953
- PMCID: PMC2757648
- DOI: 10.1038/ng.401
Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci
Abstract
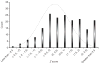
We report the results of a meta-analysis of genome-wide association scans for multiple sclerosis (MS) susceptibility that includes 2,624 subjects with MS and 7,220 control subjects. Replication in an independent set of 2,215 subjects with MS and 2,116 control subjects validates new MS susceptibility loci at TNFRSF1A (combined P = 1.59 x 10(-11)), IRF8 (P = 3.73 x 10(-9)) and CD6 (P = 3.79 x 10(-9)). TNFRSF1A harbors two independent susceptibility alleles: rs1800693 is a common variant with modest effect (odds ratio = 1.2), whereas rs4149584 is a nonsynonymous coding polymorphism of low frequency but with stronger effect (allele frequency = 0.02; odds ratio = 1.6). We also report that the susceptibility allele near IRF8, which encodes a transcription factor known to function in type I interferon signaling, is associated with higher mRNA expression of interferon-response pathway genes in subjects with MS.
Figures
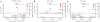

References
-
- Hauser SL, Oksenberg JR. The neurobiology of multiple sclerosis: genes, inflammation, and neurodegeneration. Neuron. 2006;52:61–76. - PubMed
-
- Barcellos LF, et al. Heterogeneity at the HLA-DRB1 locus and risk for multiple sclerosis. Hum Mol Genet. 2006;15:2813–2824. - PubMed
-
- Hafler DA, et al. Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med. 2007;357:851–862. - PubMed
-
- Rubio JP, et al. Replication of KIAA0350, IL2RA, RPL5 and CD58 as multiple sclerosis susceptibility genes in Australians. Genes Immun. 2008;9:624–630. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

